- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 14
Biochemistry
Ubiquitination Assay for Mammalian Cells
Determination of Intra- and Extracellular Glucose in Mycelium of Fusarium oxysporum
Determination of Intracellular ATP Levels in Mycelium of Fusarium oxysporum
Investigating the Assembly Status of the Plastid Encoded Polymerase Using BN-PAGE and Sucrose Gradient Centrifugation
Expression, Purification and Crystallization of the Herpesvirus Nuclear Egress Complex (NEC)
Developmental Biology
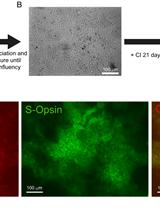
Differentiation of Human Embryonic Stem Cells into Cone Photoreceptors
Immunology
House Dust Mite Extract and Cytokine Instillation of Mouse Airways and Subsequent Cellular Analysis
Microbiology
Extraction and Quantification of Polyphosphate in the Budding Yeast Saccharomyces cerevisiae
A Bioassay Protocol for Quorum Sensing Studies Using Vibrio campbellii
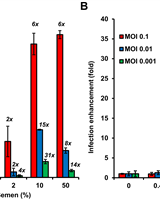
Reporter Assay for Semen-mediated Enhancement of HIV-1 Infection
Fluorescent Detection of Intracellular Nitric Oxide in Staphylococcus aureus
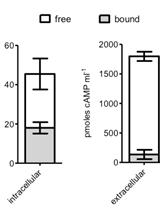
Separation of Free and Bound cAMP in Mycobacteria
Neuroscience
Mouse Subependymal Zone Explants Cultured on Primary Astrocytes
Plant Science
Isolation of Flavonoids from Piper delineatum Leaves by Chromatographic Techniques
Establishment of a Fusarium graminearum Infection Model in Arabidopsis thaliana Leaves and Floral Tissues