- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 24
Biochemistry
Activity-based Pull-down of Proteolytic Standard and Immunoproteasome Subunits
Cancer Biology
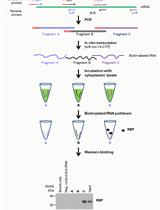
Affinity Pulldown of Biotinylated RNA for Detection of Protein-RNA Complexes
In vitro Assays for the Detection of Calreticulin Exposure, ATP and HMGB1 Release upon Cell Death
Cell Biology
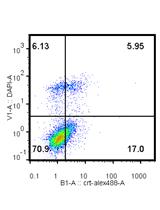
Isolation of THY1+ Undifferentiated Spermatogonia from Mouse Postnatal Testes Using Magnetic-activated Cell Sorting (MACS)
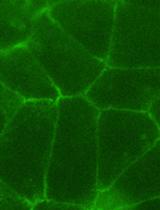
Measurement of Mechanical Tension at cell-cell junctions using two-photon laser ablation
Microbiology
Antibiotic Disc Assay for Synechocystis sp. PCC6803
Highly Accurate Real-time Measurement of Rapid Hydrogen-peroxide Dynamics in Fungi
Single-step Marker Switching in Schizosaccharomyces pombe Using a Lithium Acetate Transformation Protocol
Molecular Biology
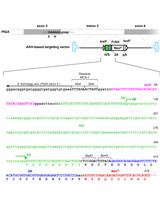
Efficient AAV-mediated Gene Targeting Using 2A-based Promoter-trap System
Plant Science
Plant Tissue Trypan Blue Staining During Phytopathogen Infection
Detection of Reactive Oxygen Species in Oryza sativa L. (Rice)
Infection of Nicotiana benthamiana Plants with Potato Virus X (PVX)
Inoculation of Rice with Different Pathogens: Sheath Blight (Rhizoctonia solani), Damping off Disease (Pythium graminicola) and Barley Powdery Mildew (Blumeria graminis f. sp. hordei)
Assessment of Wheat Resistance to Fusarium graminearum by Automated Image Analysis of Detached Leaves Assay
Electro-fusion of Gametes and Subsequent Culture of Zygotes in Rice
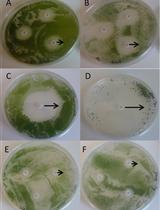
Bacterial Growth Inhibition Assay for Xanthomonas oryzae pv. oryzae or Escherichia coli K12 Grown together with Plant Leaf Extracts
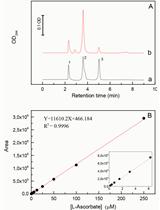
Quantitative Determination of Ascorbate from the Green Alga Chlamydomonas reinhardtii by HPLC
Expression, Purification and Crystallization of Recombinant Arabidopsis Monogalactosyldiacylglycerol Synthase (MGD1)
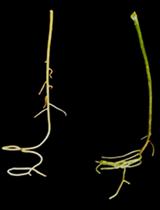
Protocol for Increasing Carotenoid Levels in the Roots of Citrus Plants
Stem Cell
Gene Expression Analysis of Sorted Cells by RNA-seq in Drosophila Intestine
Quantitative 3D Time Lapse Imaging of Muscle Progenitors in Skeletal Muscle of Live Mice