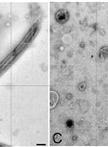
Ciliary and Flagellar Membrane Vesicle (Ectosome) Purification
Eukaryotic cilia/flagella are ideal organelles for the analysis of membrane trafficking, membrane assembly, and the functions of a variety of signal transduction molecules. Cilia are peninsular organelles and the membrane lipids, membrane proteins, and microtubular-associated components are selectively transported into cilia through the region formed by the basal body/transition region and tightly associated ciliary membrane. Cilia can be isolated from many organisms without disrupting cells and many will rapidly regenerate cilia (with the ciliary membrane lipids and proteins) to replace those that are released. Despite their ease of isolation, we have relatively little understanding of the mechanisms that regulate lipid and protein transport into ciliary membranes (Pazour and Bloodgood, 2008; Bloodgood, 2009; Bloodgood, 2012).Chlamydomonas flagella shed membrane vesicles, also called ectosomes (Wood et al., 2013) from flagellar tips and these vesicles can be purified from the culture medium without damaging or deflagellating cells (McLean et al., 1974; Bergman et al., 1975; Snell, 1976; Kalshoven et al., 1990). Based on a comparison of biotinylated proteins on the shed vesicles with biotinylated proteins isolated from purified flagella and cell bodies, the ectosomes contain most, but not all, flagellar surface proteins and none of the major cell body proteins (Dentler, 2013). Although ectosomes have only been purified from Chlamydomonas cells, preliminary evidence indicates that similar vesicles are released from Tetrahymena cilia (Dentler, unpublished). Flagellar (and ciliary) membranes or membrane proteins also can be released from purified flagella/cilia. Most membrane proteins can be solubilized by extracting purified cilia with nonionic detergent [Triton X-100 or X-114 or Nonidet P-40 (NP-40)] and pelleting the microtubules (axonemes). However, not all membranes are released by detergent (Dentler, 1980) and the supernatant also contains all of the flagellar proteins that are not attached to the microtubules. Intact membrane vesicles can be released from flagella by agitation of flagella, often with low concentrations of nonionic detergents or freeze-thawing (Witman et al., 1972; Snell, 1976; Dentler, 1980; Dentler, 1995; Bloodgood and May, 1982; Pasquale and Goodenough, 1987; Iomini et al., 2006; Huang et al., 2007). Once released, they can be purified from axonemes by differential centrifugation.Each of these methods may enrich for different populations of axonemal and membrane proteins and lipids. The different solubility of membranes may reveal local differences in lipid or protein composition (Bloodgood, 2009). The ectosomes contain most but not all surface proteins found on purified Chlamydomonas flagella (Dentler, 2013). The ectosomes vesicles may be enriched in different soluble flagellar proteins than those trapped as vesicles are released from purified flagella. The detergent-solubilized “membrane+matrix” will contain all soluble membrane proteins as well as all of the soluble proteins in the flagellar compartment. In this paper, a method to purify ectosomes vesicles released from the tips of living Chlamydomonas cells is presented as are two methods to release flagellar membrane vesicles and proteins from purified flagella.