- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rhizosphere Acidification Assay
Published: Vol 5, Iss 23, Dec 5, 2015 DOI: 10.21769/BioProtoc.1676 Views: 7610
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Experiments for in silico evaluation of Optimality of Photosynthetic Nitrogen Distribution and Partitioning in the Canopy: an Example Using Greenhouse Cucumber Plants
Yi-Chen Pao [...] Hartmut Stützel
Mar 20, 2020 4361 Views

Accelerated Snowmelt Protocol to Simulate Climate Change Induced Impacts on Snowpack Dependent Ecosystems
Laura T. Leonard [...] Jonathan O. Sharp
Mar 20, 2020 3383 Views

Quantification of Ethylene Production in Leaf and Bud Tissue of the Subtropical Tree Crop Litchi (Litchi chinensis Sonn.) Using Gas Chromatography and Flame Ionization Detection
Regina B. Cronje and Arnoldus J. Jonker
Mar 20, 2023 421 Views
Abstract
Plant survival depends on the ability of root systems to establish themselves in locations where water and nutrients are available for uptake and translocation (Hawes et al., 2003). Rhizosphere influences crop productivity by mediating efficient nutrient transformation, acquisition, and use (Shen et al., 2013). Rhizosphere acidification is a central mechanism for plant mineral nutrition since it contributes to nutrient solubility and the proton motive force (pmf). This pmf is generated by the plasma membrane H+-ATPases (Miller and Smith, 1996; Forde, 2000) in root epidermal and cortical cells, and is coupled to active nutrient acquisition (e.g., N, K, P). Roots are able to acidify the rhizosphere by up to two pH units compared to the surrounding bulk soil mainly through the release of protons, but also bicarbonate, organic acids and CO2. Here we present an easy and inexpensive protocol to quantify protons released to the media by the root system-a method successfully used in our recently published work (Pizzio et al., 2015).
Keywords: RootMaterials and Reagents
- Arabidopsis thaliana seeds (Col-0)
- Eppendorf tubes (1.5 ml)
- Commercial Bleach
- Pipettes and tips (1 and 5 ml)
- Silicon caps (or aluminum foil)
- Square Petri dishes
- Plastic wrap
- Spatula and forceps
- Flasks (200 ml)
- Plastic wrap
- Glass culture tubes
- Tween-20 (Sigma-Aldrich, catalog number: P-1379 )
- Murashige and Skoog medium (MS) (PhytoTechnology Laboratories®, catalog number: M524 )
- Sucrose (VWR International, catalog number: BDH-0308 )
- Potassium Hydroxide (KOH) (Thermo Fisher Scientific, catalog number: P-250 )
- Agar-agar (Sigma-Aldrich, catalog number: A-1296 )
- Sterile distilled water
- MES hydrate (Sigma-Aldrich, catalog number: M-8250 )
- Seed sterilization solution (see Recipes)
- MS solid (see Recipes)
- MS liquid (see Recipes)
- Assay solution (see Recipes)
Equipment
- Autoclave
- Rotary shaker
- Flow hood
- Fridge (4 °C)
- Plant growth chamber (Conviron, catalog number: ATC26 )
- Magnetic stirrer and stirring bars
- pH meter (Beckman Coulter)
Note: pH probe should be capable of measuring pH in samples with volumes <= 3 ml.
- Balance
Procedure
- Seed sterilization
- Put 5 mg (200-300 seeds) of Arabidopsis seeds into an Eppendorf tube and add 750 µl sterilization solution.
- Vortex for 15 min at room temperature.
- Decant the sterilization solution (with sterile tips) under flow hood and add 750 µl sterile water.
- Vortex briefly.
- Repeat steps c and d 3 times.
- Stratify the seeds in 750 µl sterile water at 4 °C for 2 d in the dark.
- Put 5 mg (200-300 seeds) of Arabidopsis seeds into an Eppendorf tube and add 750 µl sterilization solution.
- Plating seeds (under flow hood)
- Sow seeds (about 10) on sterile solid MS plates. Keep a density of approximately one seed per cm2. Crowded plates produce unhealthy seedlings.
- Close and seal plates with plastic wrap. Use only one layer of plastic wrap. More than one layer prevents gaseous exchange and healthy plant growth.
- Incubate plates in a vertical position in a growth chamber at 21 °C with a 12-h-light/12-h-dark cycle (150 μmol m-2 s-1) (Forde, 2000). Vertical growth reduces root stress and prevents agar transfer when transplanting seedlings.
- Grow seedlings for 5-7 d.
- Sow seeds (about 10) on sterile solid MS plates. Keep a density of approximately one seed per cm2. Crowded plates produce unhealthy seedlings.
- Plant growth in liquid media
- Prepare 200 ml flasks with 4 ml liquid MS each.
- Close flasks with a “silicon” cap (or with aluminum foil).
- Autoclave flasks at 121 °C for 20 min.
- Let flasks cool in flow hood.
- Transfer 10 seedlings into each flask. Put seedlings in the bottom with the help of sterile spatula and forceps. Be sure that the seedlings stay in contact with the media.
- Close flasks with a layer of plastic wrap. Transparent plastic wrap favors transmission of light, promotes uniform plant growth, and prevents media evaporation.
- Grow plants on a rotary shaker at 50 rpm in chamber at 21 °C with a 12-h-light/12-h-dark cycle (150 μmol m-2 s-1) for 2 weeks.
- Pay attention: usually seedlings consume all the liquid media before the 2-week period. In this case, add 4 ml sterile liquid MS.
- Prepare 200 ml flasks with 4 ml liquid MS each.
- Acidification assay
- Empty flasks and wash roots with the assay solution (5 ml) for 5 min.
- Empty flasks again and add fresh assay solution (3 ml).
- Prepare a control flask with assay solution (3 ml), but without any plants.
- Close flasks with plastic (transparent) foil and keep them in the growth chamber for 6 h.
- Transfer all the 3 ml of assay solution from flasks to a glass tube and proceed to pH measurements.
- Take out plants from flasks and check the number of plants in each flask.
- Dry plants with paper towels, cut roots from shoots, and immediately weigh to avoid plant dehydration, and the concomitant weight loss. Root and shoot weights are important values because we can either calculate protons released per gram of root or seedling.
- Empty flasks and wash roots with the assay solution (5 ml) for 5 min.
- Calculation
- Use pH values from each flask to compute [H+] (mole/L) in sample and control using the equation pH=-log [H+].
- Calculate the number of H+ moles in 3 ml of experimental and control samples [mole/ 3 ml].
- Compute crude H+ released to the medium by subtracting the average values of H+control from H+sample. A typical experiment requires from 3 to 5 independent replicates.
- Relativize average number of H+ moles to either plant number, or to root or plant fresh weight. The units are going to be either mole/plant or mole/g. These relativized values allow us to calculate an average from several flasks and also to compare flasks with different plant numbers or plant lines.
- Use pH values from each flask to compute [H+] (mole/L) in sample and control using the equation pH=-log [H+].
Representative data
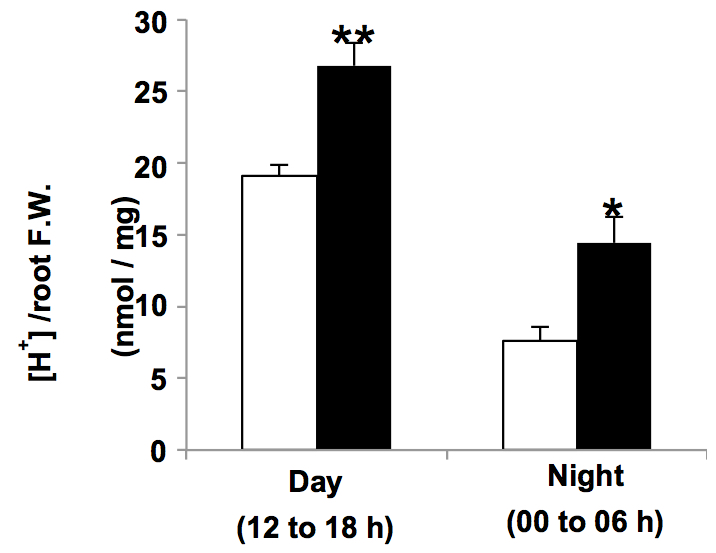
Figure 1. Protons released from the roots during day and night hours by Col-0 (empty bars) and transgenic plants overexpressing the type I H+-PPase AVP1 (AVP1-1; Pizzio et al., 2015) (black bars) plants grown in liquid media (mean ± SE; n= 6 pools of 10 plants per line, per time of day, per trial; two independent trials)
Recipes
- Seed sterilization solution
30% (v/v) commercial bleach
0.05% (v/v) Tween-20
- Murashige and Skoog (MS) solid
One-half-strength MS
1% [w/v] Sucrose
pH 5.7 with KOH
1% [w/v] agar
- MS liquid
One-half-strength MS+
1% [w/v] Sucrose (pH 5.7 with KOH)
- Assay solution
One-quarter-strength MS
2 mM MES buffer (pH 6.8 with KOH). Low MES concentration assures similar starting pH for all plants.
Acknowledgments
RAG, GAP, and KR were supported by the National Science Foundation (grant no. IOS-1122148). This protocol was adapted from Pizzio et al. (2015).
References
- Forde, B. G. (2000). Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465(1-2): 219-235.
- Hawes, M. C., Bengough, G., Cassab, G. and Ponce, G. (2003). Root caps and rhizosphere. J Plant Growth Regul 21: 352-367.
- Miller, A. J. and Smith, S. J. (1996). Nitrate transport and compartmentation in cereal root cells. Journal of Experimental Botany, Vol. 47, No. 300, pp. 843-854.
- Pizzio, G. A., Paez-Valencia, J., Khadilkar, A. S., Regmi, K., Patron-Soberano, A., Zhang, S., Sanchez-Lares, J., Furstenau, T., Li, J., Sanchez-Gomez, C., Valencia-Mayoral, P., Yadav, U. P., Ayre, B. G. and Gaxiola, R. A. (2015). Arabidopsis type I proton-pumping pyrophosphatase expresses strongly in phloem, where it is required for pyrophosphate metabolism and photosynthate partitioning. Plant Physiol 167(4): 1541-1553.
- Shen, J., Li, C., Mi, G., Li, L., Yuan, L., Jiang, R. and Zhang, F. (2013). Maximizing root/rhizosphere efficiency to improve crop productivity and nutrient use efficiency in intensive agriculture of China. J Exp Bot 64(5): 1181-1192.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Pizzio, G. A., Regmi, K. and Gaxiola, R. (2015). Rhizosphere Acidification Assay. Bio-protocol 5(23): e1676. DOI: 10.21769/BioProtoc.1676.
-
Pizzio, G. A., Paez-Valencia, J., Khadilkar, A. S., Regmi, K., Patron-Soberano, A., Zhang, S., Sanchez-Lares, J., Furstenau, T., Li, J., Sanchez-Gomez, C., Valencia-Mayoral, P., Yadav, U. P., Ayre, B. G. and Gaxiola, R. A. (2015). Arabidopsis type I proton-pumping pyrophosphatase expresses strongly in phloem, where it is required for pyrophosphate metabolism and photosynthate partitioning. Plant Physiol 167(4): 1541-1553.
Category
Plant Science > Plant physiology > Plant growth
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








