- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Lentiviral Knockdown of Transcription Factor STAT1 in Peromyscus leucopus to Assess Its Role in the Restriction of Tick-borne Flaviviruses
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2643 Views: 9600
Reviewed by: Emilie BesnardFei XiaoJose Antonio Reyes-Darias

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Isolation and Culture of Primary Fibroblasts from Neonatal Murine Hearts to Study Cardiac Fibrosis
Shweta Kumar [...] Nagalingam Ravi Sundaresan
Feb 20, 2023 1023 Views
Protein Pull-down Assay Using HiBiT-tag-dependent Luciferase Activity Measurement
Masashi Arakawa and Eiji Morita
Mar 20, 2023 716 Views
Methods to Quantify the Dynamic Recycling of Plasma Membrane Channels
Rawad Hodeify and Khaled Machaca
Sep 5, 2023 510 Views
Abstract
Cellular infection with tick-borne flaviviruses (TBFVs) results in activation of the interferon (IFN) signaling pathway and subsequent upregulation of numerous genes termed IFN stimulated genes (ISGs) (Schoggins et al., 2011). Many ISGs function to prevent virus pathogenesis by acting in a broad or specific manner through protein-protein interactions (Duggal and Emerman, 2012). The potency of the IFN signaling response determines the outcome of TBFV infection (Best, 2017; Carletti et al., 2017). Interestingly, data from our lab show that TBFV replication is significantly restricted in cells of the reservoir species Peromyscus leucopus thereby suggesting a potent antiviral response (Izuogu et al., 2017). We assessed the relative contribution of IFN signaling to resistance in P. leucopus by knocking down a major transcription factor in the IFN response pathway. Signal transducer and activator of transcription 1 (STAT1) was specifically targeted in P. leucopus cells by shRNA technology. We further tested the impact of gene knockdown on the ability of cells to respond to IFN and restrict virus replication; the results indicate that when STAT1 expression is altered, P. leucopus cells have a decreased response to IFN stimulation and are significantly more susceptible to TBFV replication.
Keywords: InterferonBackground
IFN signaling is the first line of defense against flaviviruses invading a host cell (Robertson et al., 2009; Lazear and Diamond, 2015). Molecular signatures associated with the virus particles are detected by pattern-recognition receptors (PRRs) which then elicit downstream signaling via transcription factors to release type 1 IFNs from the cell (Kawai and Akira, 2011; Loo and Gale, 2011). Further, the IFN response pathway is initiated by binding of IFN-beta (IFN-β) to its cognate receptor and activation of the JAK-STAT signaling cascade (Loo et al., 2008). STAT1 is a major player in the IFN response pathway and functions within a complex to activate the IFN–stimulated response element (ISRE) promoter in the nucleus (Stark et al., 1998). Ultimately, IFN signaling transcriptionally upregulates numerous host genes that act to curtail infection and limit viral pathogenesis (Liu et al., 2011).
TBFVs contribute to the global burden of disease by causing encephalitis or hemorrhagic fevers in infected individuals. There are about 10,000 cases of TBE annually and disease is endemic to Europe and parts of Asia (Suss, 2008). In recent years, TBFVs have shown emerging nature due to their occurrence in previously unreported regions and re-occurrence in areas of eradication (Robertson et al., 2009; Ebel, 2010). The reason for the increased spread is not known, and treatment for clinically-recognized cases is limited to palliative care (Fernandez-Garcia et al., 2009; Lani et al., 2014). While virus infection in humans and other susceptible species can result in deleterious illness, infection of a natural reservoir host P. leucopus remains asymptomatic (Telford et al., 1997; Santos et al., 2016) suggesting that the host could lack relevant proviral factors or express effective antiviral factors. Data from our lab has explored these possibilities and demonstrated that viral restriction in P. leucopus cells is mediated by an antiviral response occurring via the IFN signaling cascade (Izuogu et al., 2017). Our studies involved resolving the sequence of antiviral gene homologs in P. leucopus and targeting components of the IFN response pathway. Specifically, our study first showed that STAT1 expression was consistently higher in P. leucopus cells compared to the susceptible control M. musculus following IFN treatment and virus infection (Izuogu et al., 2017). Based on these data, we further assessed the relative role of STAT1 in virus restriction by specific gene knockdown. This protocol describes the technique used to design shRNA to target STAT1, packaging into lentiviruses and cellular transduction to establish P. leucopus stable cell lines with diminished STAT1 expression. We further provide details of how these cells were assayed for a loss of restriction following infection with a TBFV, Langat virus (LGTV).
Materials and Reagents
- Materials
- Tissue culture plates (Greiner Bio One International, catalog numbers: 657160 , 662160 , 677180 –6, 24, and 48 wells respectively)
- Tissue culture flasks (Greiner Bio One International, catalog numbers: 658175 and 660175 –75 cm2 and 175 cm2 respectively)
- Eppendorf tubes (1.5 ml) (USA Scientific, catalog number: 1615-5510 )
- PVDF membranes (Thermo Fisher Scientific, catalog number: 88518 )
- Lightcycler 480 96-well plate (Roche Molecular Systems, catalog number: 04729692001 )
- Labtek slides (Thermo Fisher Scientific, catalog number: 154534 )
- 1.2 ml tubes (USA Scientific, catalog number: 1412-1000 )
- Autoradiography film (McKesson Medical-Surgical, catalog number: EBA45 )
- Coverslips (Fisher Scientific, catalog number: 12-545-88 )
- Reagent reservoir troughs (VistaLab, catalog number: 3054-1001 )
- Cell scraper (Fisher Scientific, catalog number: 08-100-241 )
- Airtight container (Sterilite, catalog number: 1642 )
- XCell4 SureLock Midi SDS-PAGE system (Thermo Fisher Scientific, InvitrogenTM, catalog number: WR0100 )
- 20-well Tris-Glycine midi gel (Thermo Fisher Scientific, InvitrogenTM, catalog number: WT0102BOX )
- Tissue culture plates (Greiner Bio One International, catalog numbers: 657160 , 662160 , 677180 –6, 24, and 48 wells respectively)
- Cells
- Human embryonic kidney (HEK)-293T cells (ATCC, catalog number: CRL-3216 )
- HEK-293 cells (ATCC, catalog number: CRL-1573 )
- African green monkey kidney cells–Vero cells (ATCC, catalog number: CCL-81 )
- P. leucopus adult skin fibroblast cells (Coriell Institute, catalog number: AG22353 )
- One ShotTM TOP10 Chemically-Competent E. coli cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: C404010 )
- Human embryonic kidney (HEK)-293T cells (ATCC, catalog number: CRL-3216 )
- Viruses
Langat virus (LGTV) strain TP21 (kindly provided by Dr. Sonja M. Best, NIAID/NIH)
Note: LGTV is a naturally-attenuated member of the tick-borne flavivirus serocomplex used under biosafety level 2 (BSL2) laboratory conditions. All safety precautions for BSL2 level containment were adhered to. - Reagents
- pLENTI X2- DEST vector (Campeau et al., 2009)
- Recombinant mouse IFN-β (Pestka Biomedical Laboratories, catalog number: 12405-1 )
- Oligonucleotide primer pairs (see Table 1)
- RNAi BLOCK-iT U6 RNAi Entry Vector Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: K4945-00 )
- Agarose (BioExpress,, catalog number: E-3120-500 )
- 100 bp DNA ladder (New England Biolabs, catalog number: N3231L )
- TAE (10x) buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15558042 )
- 10x Tris-glycine SDS-PAGE running buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: LC2675 )
- LR Clonase II Enzyme Mix (Thermo Fisher Scientific, catalog number: 11791100 )
- ViraPowerTM Lentiviral Packaging Mix (Thermo Fisher Scientific, catalog number: K497500 )
- Lipofectamine 3000 (Thermo Fisher Scientific, InvitrogenTM, catalog number: L3000015 )
- Sodium butyrate (Sigma-Aldrich, catalog number: B5887 )
- Lenti-X GoStix (Takara Bio, Clontech, catalog number: 631244 )
- Polybrene (Sigma-Aldrich, catalog number: 107689 )
- Hygromycin B (Thermo Fisher Scientific, GibcoTM, catalog number: 10687010 )
- Phosphate buffered saline (PBS) (Thermo Fisher Scientific, InvitrogenTM, catalog number: QVC0508 ) alone or with 0.5% Tween-20 (PBST)
- iBlot transfer stacks (Thermo Fisher Scientific, InvitrogenTM, catalog number: IB4010-01 )
- Antibodies
- α-actin (Sigma-Aldrich, catalog number: A5441 )
- α-STAT1 (Cell Signaling Technology, catalog number: 9172S , used at 1:1,000) detects phosphorylation of STAT1 at Tyr701
- α-STAT1-P (Cell Signaling Technology, catalog number: 9167S , used at 1:1,000)
- α-LGTV E (provided by Dr. C. Schmaljohn, USAMRIID used at 1:1,000)
- α-LGTV NS3 (as described in Taylor et al., 2011, used at 1:2,000)
- Goat anti-mouse IgG (Thermo Fisher Scientific, catalog number: A28177 , used at 1:3,000 for immunoblotting and at 1:1,000 viral for immunofocus assays)
- Rabbit anti-chicken AlexaFluor 594 (Thermo Fisher Scientific, catalog number: A11042 , used at 1:1,000)
- Goat anti-mouse AlexaFluor 488 (Thermo Fisher Scientific, catalog number: A11029 , used at 1:1,000)
- α-actin (Sigma-Aldrich, catalog number: A5441 )
- ECL Plus (Thermo Fisher Scientific, PierceTM, catalog number: 80196 )
- RNeasy Mini Kit (QIAGEN, catalog number: 74104 )
- QuantiTect Reverse Transcription Kit (QIAGEN, catalog number: 205310 )
- PCR grade water (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9935 )
- Gene-specific qRT-PCR primers (see Table 2)
- FastStart essential DNA Green master (Roche Molecular Systems, catalog number: 06402712001 )
- Paraformaldehyde 16% w/v (Alfa Aesar, catalog number: 43368 )
- ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36931 )
- Methanol
- Opti-MEM reduced serum medium (Thermo Fisher Scientific, catalog number: 31985062 )
- Tris (Fisher Scientific, catalog number: BP152-5 )
- Sodium chloride (NaCl) (Gentrox, catalog number: 60-037 )
- Deoxycholate (Sigma-Aldrich, catalog number: D6750 )
- 10x Tris-glycine buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: LC2672 )
- Sodium dodecyl sulfate (SDS) (Fisher Scientific, catalog number: BP166-500 )
- NP-40 (IGEPAL CA-630) (Sigma-Aldrich, catalog number: 542334 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Avantor Performance Materials, MACRON, catalog number: 5958 )
- Calcium chloride (CaCl2) (Acros Organics, catalog number: 192735000 )
- 3,3-Diaminobenzidine HCl (Sigma-Aldrich, catalog number: D5637 )
- Hydrogen peroxide (H2O2) (Sigma-Aldrich, catalog number: H3410 )
- Glycerol (AMRESCO, catalog number: 0854 )
- Bromophenol blue (Bio-Rad Laboratories, catalog number: 1610404 )
- β-Mercaptoethanol (Alfa Aesar, catalog number: J61337 )
- Non-fat dry milk
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: 12100061 )
- Methylcellulose (Fisher Scientific, catalog number: M352-500 )
- Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11550 )
- Triton X-100 ( Sigma Aldrich, catalog number: X100 )
- Sodium citrate (Sigma-Aldrich, catalog number: PHR1416 )
- Bovine serum albumin (BSA)
- Goat serum
- Quibit DNA BR kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32850 )
- Quibit RNA HS kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32852 )
- RNase-free DNase set (QIAGEN, catalog number: 79254 )
- Complete EDTA-free protease inhibitor cocktail (Thermo Fisher Scientific, PierceTM, catalog number: A32955 )
- Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics, catalog number: 11873580001 )
- DC Protein assay reagents (Bio-Rad Laboratories, catalog number: 5000116 )
- Radioimmunoprecipitation assay (RIPA) buffer (see Recipes)
- DNase I buffer (10x) (see Recipes)
- Peroxidase substrate (prepared fresh) (see Recipes)
- 6x SDS-PAGE sample buffer (6x SB) (see Recipes)
- Western blot blocking solution (see Recipes)
- Culture overlay media (see Recipes)
- Permeabilization solution (see Recipes)
- Immunofluorescence assay blocking solution (see Recipes)
- pLENTI X2- DEST vector (Campeau et al., 2009)
Equipment
- Pipettes (USA Scientific)
- Transilluminator (Omega Ultra-Lum)
- Tabletop centrifuge (Eppendorf)
- iBlot dry blotting transfer machine (Thermo Fisher Scientific)
- Rocker (Benchmark)
- Rotator (Bellco Biotechnology)
- Autoradiography film processor (Kodak)
- Thermocycler (Roche Lightcycler)
- Tissue culture incubator (at 37 °C with 5% CO2) (Thermo Fisher Scientific)
- Biosafety cabinet
- Water bath (VWR Scientific)
- Heating block (at 95 °C) (Fisher Scientific)
- SDS-PAGE apparatus (Pharmacia Biotech)
- Refrigerator
- Vortex (Fisher Scientific)
- Qubit 2.0 fluorometer (Thermo Fisher Scientific, current model: Qubit 3.0 )
- Confocal microscope (Olympus Life Science)
Software
- GraphPad Prism 6: https://www.graphpad.com/scientific-software/prism/
- Roche Lightcycler 96 software: https://lifescience.roche.com/en_us/brands/realtime-pcr
- Invitrogen primer design software: https://rnaidesigner.thermofisher.com/rnaiexpress/
- MacVector software version 12.7: http://macvector.com/EcoRI/macvector12.7.5installer.html
Procedure
- Design and cloning of short hairpin RNA (shRNA) into lentiviral vectors
Our group recently resolved and published the sequence of STAT1 in the reservoir species P. leucopus (Izuogu et al., 2017) with accession number KY451962. Based on the gene sequence, the oligonucleotides for knockdown were designed and cloned as described below; A flowchart describing the process of oligonucleotide processing and generation of lentiviral vectors is represented in Figure 1.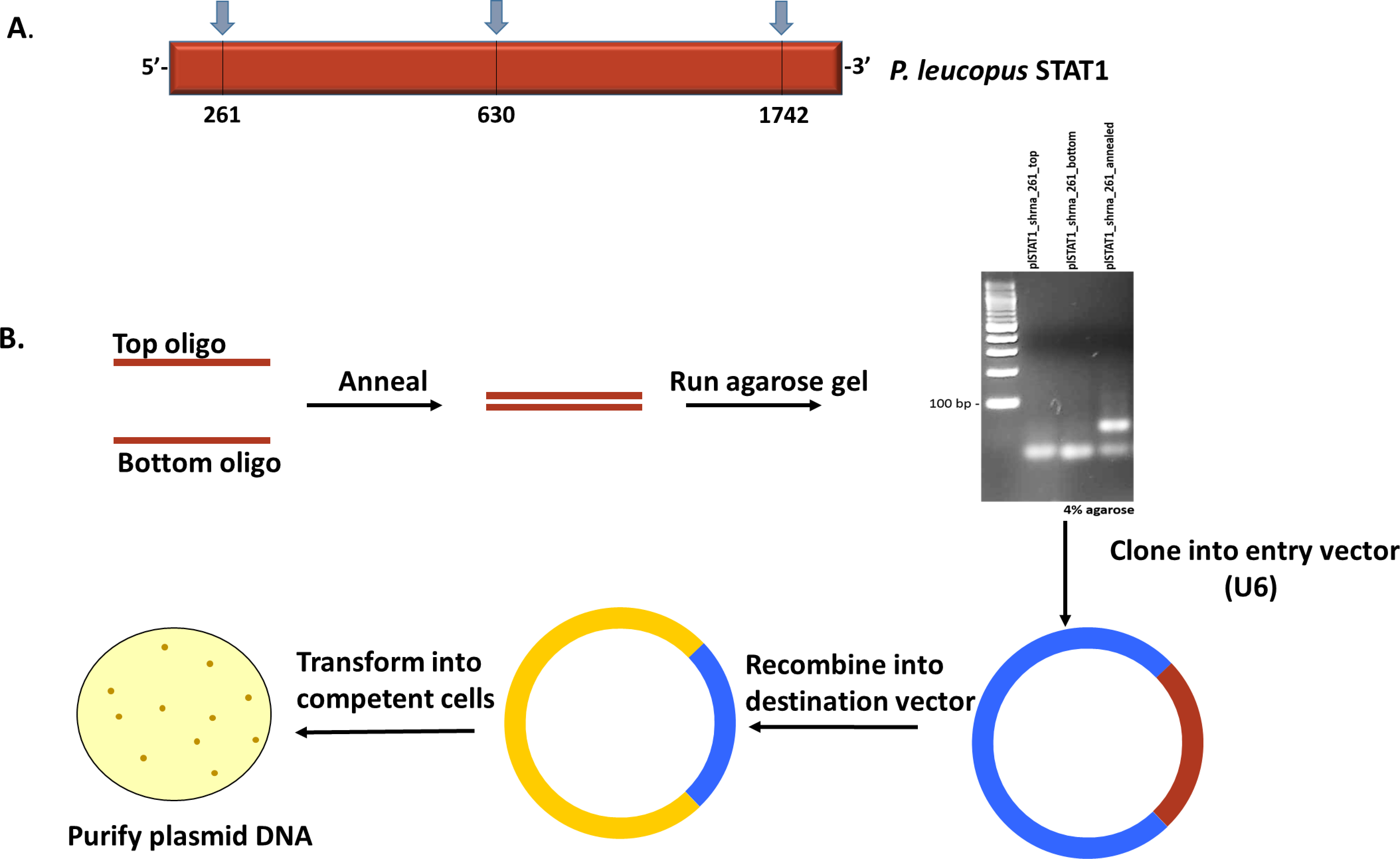
Figure 1. Construction of lentiviral plasmids. A. Schematic representation of P. leucopus STAT1 showing the regions targeted for knockdown. B. Workflow of procedure to generate lentiviral expression vectors with shRNA to P. leucopus STAT1.- Upload the complete sequence to the Invitrogen primer design software (or equivalent) and generate oligo candidates for shRNA design. Regions around the beginning, middle and end of the gene are targeted to ensure complete transcript coverage and avoid off-target effects. Candidates selected are shown in Table 1 and Figure 1A.
Table 1. Oligonucleotide sequences utilized for gene knockdown. shRNA top and bottom strand sequences designed to target 3 distinct regions of the P. leucopus STAT1 gene. No special conditions were specified in the primer order.
- As a control, design non-specific oligonucleotides to create lentiviral vectors that will not result in knockdown. An shRNA that targets the GFP sequence was used for this purpose in our study (see Table 1). Since GFP is not expressed in the cells of interest, the control cell lines should be transcriptionally equivalent to the background, non-transduced cells.
- Anneal the top and bottom strands by mixing oligonucleotides in equal proportion (200 μM each) and subjecting to high temperature (95 °C) for 4 min according to the manufacturer’s annealing protocol for the BLOCK-iT U6 RNAi Entry Vector Kit (Thermo Fisher scientific). After the heating process, immediately transfer the samples to the bench and allow for cooling to occur slowly. This process will generate double-strand shRNA.
- To confirm the annealing step, resolve the samples on a 4% agarose gel (immersed in 1x TAE buffer) alongside a 100 bp ladder by adding equal volume of each sample (10 μl) to single wells and running at 100 V. Transfer the gel to a transilluminator to visualize double-strand oligonucleotides compared to the single-strand samples (Illustrated in Figure 1B).
- After the double-strand is confirmed by agarose gel electrophoresis, proceed with the annealed samples and ligate the resultant shRNA into the U6 vector to generate an entry vector for further cloning. The U6 vector is contained within the BLOCK-iT U6 RNAi Entry Vector Kit (Thermo Fisher Scientific) and all manufacturers’ recommendations were adhered to.
- Recombine the entry vector into a lentiviral destination vector that can be delivered into mammalian cells. In this study, we utilized the pLENTI X2- DEST vector (Campeau et al., 2009) for Gateway cloning with the Gateway LR Clonase II Enzyme Mix according to the manufacturer’s protocol with the exception that recombination was allowed to proceed overnight.
- Transform chemically-competent One ShotTM TOP10 Chemically-Competent E. coli bacteria cells with the recombined DNA and select colonies for DNA extraction to obtain pure plasmid DNA.
- Upload the complete sequence to the Invitrogen primer design software (or equivalent) and generate oligo candidates for shRNA design. Regions around the beginning, middle and end of the gene are targeted to ensure complete transcript coverage and avoid off-target effects. Candidates selected are shown in Table 1 and Figure 1A.
- Generation of Lentiviruses
- Set up 293T cells at 3,000,000 cells per well in a 6-well dish and grow overnight.
- Transfect cells with purified lentiviral plasmids targeting the individual regions of the gene of interest and the non-specific (GFP) sequence. Importantly, include a pool of DNA containing 1 μg of lentiviral vectors targeting each region into a single transfection tube. Therefore, prepare a single tube for shRNA targeting each region of interest and an additional tube with all shRNA candidates pooled together in one tube.
- Add 1 μg of ViraPower Lentiviral Packaging Mix along with plasmid DNA and complete transfection using Lipofectamine 3000 Transfection Kit according to the manufacturer’s protocol. Pipet slowly onto cells to avoid detachment.
- At 24 h post-transfection, replace the transfection media with fresh DMEM containing 10 mM sodium butyrate, 2 ml total volume was suitable for a 6-well dish in our studies.
- Incubate the cells for another 48 h without perturbing the culture dishes.
- Collect supernatants containing the newly-generated lentiviruses. Confirm the presence of lentiviruses by running a rapid test using Lenti-X GoStix (use 20 µl sample supernatant followed by three drops of the kit running buffer and allow time for bands to develop). Lentiviral supernatants were not filtered.
- Store the lentivirus stock at -80 °C until needed.
- Set up 293T cells at 3,000,000 cells per well in a 6-well dish and grow overnight.
- Transduction of P. leucopus cells with STAT1 shRNA-expressing lentiviruses
- Set up P. leucopus fibroblasts at 300,000 cells per well in 6-well dishes and incubate overnight.
- Infect the cells with 1 ml of the lentivirus stock and add 8 µg polybrene (diluted from a stock solution prepared in sterile water).
- At 1 h post-transduction, add another 1 ml of culture medium with 8 µg polybrene. 24 h later, re-transduce the cells and treat with 8 µg polybrene.
- On day 2, remove the lentiviral inoculum and replace with fresh culture medium.
- At day 3 post-transduction, add the appropriate antibiotic selection (400 µg/ml hygromycin was used for P. leucopus fibroblasts).
- Maintain the cells in selection for 1 week and sub-clone to obtain clonal cells by splitting single cells into each well of 48-well dishes and observe for cell proliferation in individual wells over 1-2 weeks.
- Expand clonal cells into large flasks (175 cm2) and proceed to test for gene knockdown.
- Set up P. leucopus fibroblasts at 300,000 cells per well in 6-well dishes and incubate overnight.
- Assessing gene knockdown in transduced cell lines
Our studies have demonstrated that STAT1 expression is upregulated in P. leucopus following treatment with IFN-β (Izuogu et al., 2017). Hence, in order to determine if the gene of interest is knocked down, the cells are treated with IFN and probed for gene induction.- Set up the STAT1 knockdown (KD) and NS KD P. leucopus fibroblasts at 1,000,000 cells per well and incubate overnight.
- Treat the cells (or not) with 1,000 international units (IU) mouse interferon beta (mIFN-β) for 8 h and harvest samples to test for knockdown as follows:
By SDS-PAGE and Western blot- Remove the cellular supernatant and wash the cells 2 x with PBS buffer.
- Add 500 µl of RIPA buffer (see Recipes) and scrape the cell monolayer gently. Pipet a few times and transfer the sample to Eppendorf tubes.
- Incubate the samples with 50 µl 1x DNase I buffer (see Recipes) for 30 min at 37 °C.
- Centrifuge at max speed for 10 min, collect supernatant and mix with 6x SB to achieve 1x SB (see Recipes). Load equal amounts of protein on 10% SDS gels and resolve at 150 V for 1.5 h.
- After the electrophoresis step, transfer the proteins to PVDF membranes using the iBLOT transfer apparatus for 7 min.
- Incubate the membranes with Western blot blocking solution (see Recipes) while rocking for 1 h at room temperature (RT).
- Probe the blots with specific antibodies to STAT1 and phosphorylated STAT1 (STAT1-P) for 16 h at 4 °C on a rocker and wash 3 x in PBST for 5 min.
- Incubate with secondary antibodies conjugated to HRP while rocking for 1 h at RT. Wash 3 x in PBST for 10 min.
- Perform a final wash with PBS for 5 min, incubate membranes with ECL-plus reagents for an additional 5 min and utilize an automated film processor to visualize protein expression. Figure 2A shows Western blot results for STAT1 KD.
By qRT-PCR- Remove cellular supernatant and wash the cells 2 x in PBS. Harvest cell lysates according to RNA extraction protocol. In this study, cells were collected in 350 µl of cell lysis buffer (RLT buffer included RNeasy mini kit) and RNA extraction was performed using the RNeasy mini kit according to the manufacturer’s protocol, including the DNase treatment step using the RNase-free DNase set (QIAGEN).
- Quantify the RNA and normalize the concentration to ensure an equal amount of RNA between all the samples.
- Perform cDNA synthesis using an equal amount of RNA template (1 µg was used). Our study utilized the QuantiTect Reverse Transcription Kit according to the manufacturer’s protocol.
- Make a 1/8 dilution of the samples using PCR-grade water and add 3 µl of each sample to individual wells of a clean, opaque 96-well plate. Add 1 µl each of Fwd. and Rev. primers for a total of 5 µl; primer sequences used to detect P. leucopus STAT1 are shown in Table 2. In this study, the FastStart Essential Green Master Enzyme Mix was used and 5 µl of this was added to each sample for the reaction. Thus, the total reaction volume was 10 µl.
Table 2. qRT-PCR primers used to probe for STAT1 and β-actin in P. leucopus cells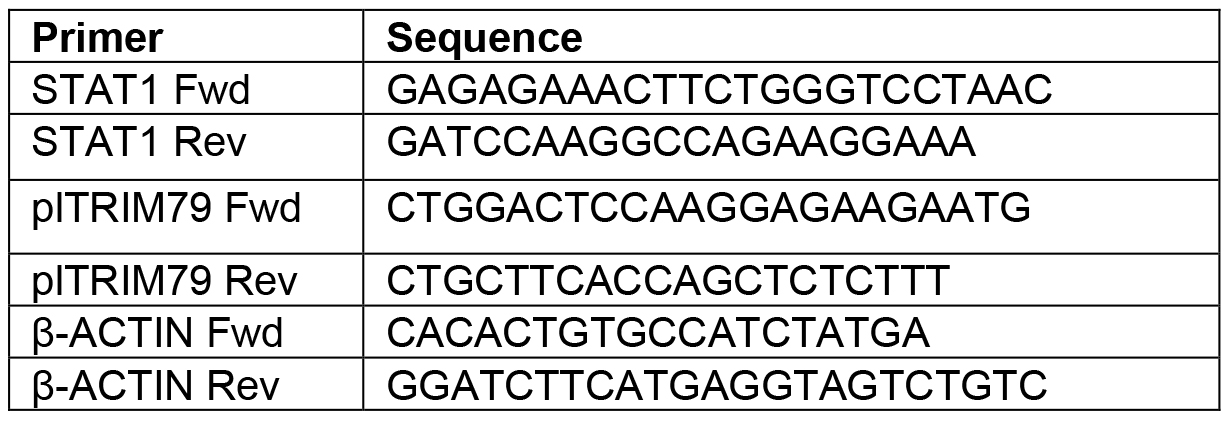
- Run on Lightcycler for 50 cycles and analyze the resultant data. The fold induction of STAT1 in the NS controls was set to 100% and the fold induction in the KD cells was assessed comparative to the NS cells. All data are normalized to β-actin. Figure 2B shows relative STAT1 mRNA expression.
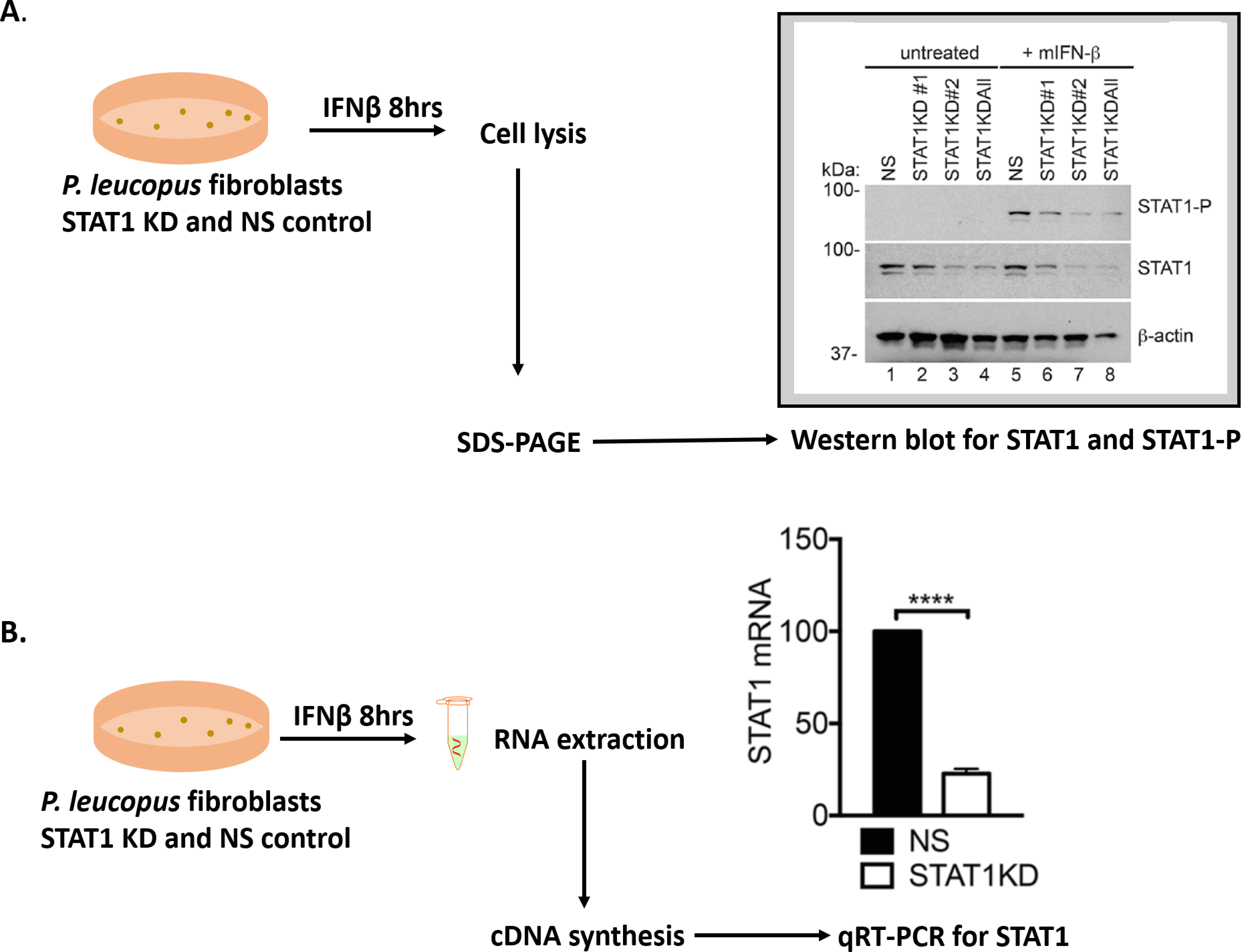
Figure 2. Assessing gene knockdown in transduced cell lines. A. Flow chart and data of Western blot showing decreased expression of STAT1 in P. leucopus KD cells expressing shRNA to positions 261 (STAT1KD #1), 630 (STAT1KD #2) and all 3 positions–261, 630 and 1,742 (STAT1KD All). All further experiments were performed with the ‘STAT1 KD All’ cells and they are henceforth named as STAT1 KD cells. Samples were probed with antibodies to STAT1, STAT1-P and β-actin. B. Flow chart and data of qRT-PCR showing percentage relative STAT1 mRNA expression in the STATIKD cells compared to the NS control. Asterisks indicate: **** = P < 0.00001.
- Remove the cellular supernatant and wash the cells 2 x with PBS buffer.
- Set up the STAT1 knockdown (KD) and NS KD P. leucopus fibroblasts at 1,000,000 cells per well and incubate overnight.
- Perform a functional assay for IFN response in STAT1 KD cells
Having observed knockdown, we tested the impact of STAT1 KD on the upregulation of an ISG. Data in our lab has identified a homolog of the TBFV restriction factor-tripartite motif protein 79 (TRIM79) and also demonstrated it to be an ISG in P. leucopus cells (Taylor et al., 2011). We assayed the fold induction of P. leucopus TRIM79 (plTRIM79) in STAT1 KD cells following IFN treatment according to the following protocol:- Set up STAT1 KD and NS control cells as in Procedure D, treat with mIFN-β for 8 h and proceed to harvest RNA as described above in the qRT-PCR section.
- Use an equal amount of RNA as template for cDNA synthesis and perform qRT-PCR reaction using gene-specific primers. Fold induction was assessed relative to untreated cells. We also compared plTRIM79 induction in P. leucopus cells with specific lentiviral knockdown to the type 1 IFN receptor (IFNAR1) generated using the same technique outlined above. Data is shown in Figure 3.
Figure 3. qRT-PCR of an ISG plTRIM79 in STAT1 KD cells relative to the NS control. Cells with altered STAT1 (and IFNAR1) expression show significantly less ISG induction suggesting that knockdown has a functional impact on IFN signaling. Mean ± SD. Asterisks indicate: ** = P < 0.01, *** = P < 0.001.
- Set up STAT1 KD and NS control cells as in Procedure D, treat with mIFN-β for 8 h and proceed to harvest RNA as described above in the qRT-PCR section.
- Test the effect of STAT1 KD on LGTV replication
By immunofluorescence microscopy- Set up STAT1 KD and NS cells at 50,000 cells per well in 8-well Labtek slides and incubate overnight.
- Infect the STAT1 KD and NS cells with LGTV at MOI of 100. Allow a 1 h absorption period and replace the inoculum with 500 µl of fresh culture media.
- At 72 h post-infection, remove supernatant, wash cells 2 x with PBS and fix for 20 min in 4% paraformaldehyde solution. Add just enough to cover the slides (about 5 ml).
- Wash 2 x with PBST and permeabilize cells for 5 min. Remove permeabilization solution (see Recipes) and add 3 ml immunofluorescence assay blocking solution (see Recipes) for 1 h at RT. Specific antibodies to LGTV envelope (E), and LGTV non-structural protein 3 (NS3) proteins were used in this study at 1:2,000 dilution.
- Wash 3 x with PBS and incubate with the appropriate fluorescent secondary antibodies diluted in 3 ml blocking solution for 1 h at RT in the dark. Wash cells 3 x in opaque containers protected from light.
- Add mounting media containing DAPI–This study utilized 1 ml of the ProLong Gold Anti-Fade Mountant with DAPI added and slides were imaged on an Olympus confocal microscope. Figure 4A includes the confocal image data showing increased viral protein staining in STAT1 KD cells.
By virus titration and immunofocus assay- Set up STAT1 KD and NS cells at 100,000 cells per well in 24-well dishes and incubate overnight.
- Infect cells with LGTV at a multiplicity of infection (MOI) of 10 such that there would be 10 virion particles to each cell in the monolayer. This is calculated as a product of the cell density, desired MOI and number of wells divided by the viral titer to determine the volume of viral stock needed for infection. Inoculate the cells with 250 µl of the virus solution and allow a 1 h absorption period and replace the inoculum with 500 µl of fresh culture media.
- Collect viral supernatants at 24, 48, and 72 h post-infection in triplicates and store at -80 °C.
- Set up Vero cells at 200,000 cells per well in 24-well dishes and incubate overnight.
- Make six 1/10 serial dilutions of each viral supernatant sample in FBS-free DMEM media using the 1.2 ml tubes and add 250 µl of each diluted to fresh Vero cells. Allow a 1 h absorption period and replace the inoculum with 500 µl of fresh culture media containing 2% FBS and 0.8% methylcellulose (culture overlay media).
- At 4 d post-infection, perform the immunofocus assay as described previously (Taylor et al., 2011; Baker et al., 2013). Briefly, remove the culture overlay media (see Recipes) and wash the cells 2 x with PBS. Fix with 100% methanol for 30 min and wash again with PBS.
- Incubate for 20 min in Opti-MEM medium and follow up with a virus-specific antibody. In this study, an antibody to detect the LGTV envelope (E) protein was used at 1:1,000. Incubate with primary antibody for 1 h at 37 °C in an airtight container.
- Wash cells 2 x with PBS and incubate with the appropriate secondary antibody. Goat anti-mouse IgG antibody was used in this study at 1:1,000. Incubate as in primary antibody and follow with 2 PBS washes.
- Add the substrate and allow 15 min for foci to develop. Determine the number of focus-forming units (FFU) per ml of viral supernatant and compare virus release in each cell type and condition. Results of STAT1 KD on LGTV infection are shown in Figure 4B.
Figure 4. Determining the impact of STAT1 KD on LGTV infection of P. leucopus. A. Visualizing LGTV protein staining in STAT1 KD P. leucopus cell lines by immunofluorescence microscopy. Images were captured on an Olympus microscope at 40x magnification showing LGTV E (green), NS3 (red) and nuclei are stained with DAPI (blue). B. Quantifying LGTV replication in P. leucopus STAT1 KD cells assessed by performing an immunofocus assay. This assay involves titrating supernatants from the infected clonal cells onto fresh Vero cells and quantifying the resultant foci. Viral titer is indicated as focus forming units (ffu) per ml. Mean ± SD; Data are from three independent experiments performed in triplicate. Asterisks indicate: * = P < 0.05, *** = P < 0.001, **** = P < 0.00001.
- Set up STAT1 KD and NS cells at 50,000 cells per well in 8-well Labtek slides and incubate overnight.
Data analysis
All titration experiments were performed three times in triplicates. Data were analyzed by an unpaired t-test or Mann-Whitney U test using GraphPad Prism 6 software as described in Izuogu et al., 2017 (http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0179781).
Notes
- Depending on the application, cDNA samples can be used at higher dilutions for qRT-PCR.
- When annealing oligonucleotides, rather than transferring the sample tubes to the bench, transfer the entire heat block from the heating system–this will allow cooling to occur at a very slow rate.
- Vero cells are African green monkey cells which have a defect in interferon secretion thereby making them suitable to count viruses by titration. They also form foci and plaques relatively well.
Recipes
- Radioimmunoprecipitation assay (RIPA) buffer
50 mM Tris pH 7.5 (100 ml 1 M)
150 mM NaCl (60 ml 5 M)
0.5% deoxycholate (20 g)
0.1% SDS (10 ml 20%)
1% NP-40 (20 ml IGEPAL CA-630)
dH2O (up to 2 L) - DNase I buffer (10x)
100 mM Tris-HCl pH 7.5
25 mM MgCl2
5 mM CaCl2 - Peroxidase substrate (prepared fresh)
0.4 mg/ml 3,3-diaminobenzidine HCl
0.45 μl/ml 30% H2O2
1x PBS - 6x SDS-PAGE sample buffer (6x SB)
375 mM Tris-HCl, pH 6.8
9% SDS
50% glycerol
0.03% bromophenol blue
9% β-mercaptoethanol, fresh - Western blot blocking solution
5% non-fat dry milk in PBST - Culture overlay media
Complete DMEM supplemented with 0.8% methylcellulose and 2% FBS - Permeabilization solution
0.1% Triton X-100 and 0.1% sodium citrate in PBS - Immunofluorescence assay blocking solution
0.5% BSA and 1% goat serum in PBS
Acknowledgments
This work is supported by a pilot grant funding from the Lyme Disease Association and the University of Toledo College of Medicine and Life Sciences startup funds (RTT). The protocol and the representative results shown herein were adapted from our published work (Taylor et al., 2011; Izuogu et al., 2017). We thank Dr. Jason Munshi-South and Dr. Stephen Harris for their help in generating the initial nucleotide hits that were used to resolve the STAT1 sequence in P. leucopus. We declare no conflicting or competing interests.
References
- Baker, D. G., Woods T. A., Butchi, N. B., Morgan, T.M., Taylor, R.T., Sunyakumthorn, P., Mukherjee, P., Lubick, K. J., Best, S. M. and Peterson K. E. (2013). Toll-like receptor 7 suppresses virus replication in neurons but does not affect viral pathogenesis in a mouse model of Langat virus infection. J Gen Virol 94(2): 336-47
- Best, S. M. (2017). The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J Virol 91(3): e01970-16.
- Campeau, E., Ruhl, V. E., Rodier, F., Smith, C. L., Rahmberg, B. L., Fuss, J. O., Campisi, J., Yaswen, P., Cooper, P. K. and Kaufman, P. D. (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4(8): e6529.
- Carletti, T., Zakaria, M. K. and Marcello, A. (2017). The host cell response to tick-borne encephalitis virus. Biochem Biophys Res Commun 492(4):533-540.
- Duggal, N. K. and Emerman, M. (2012). Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12(10): 687-695.
- Ebel, G. D. (2010). Update on Powassan virus: emergence of a North American tick-borne flavivirus. Annu Rev Entomol 55: 95-110.
- Fernandez-Garcia, M. D., Mazzon, M., Jacobs, M. and Amara, A. (2009). Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe 5(4): 318-328.
- Kawai, T. and Akira, S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34(5): 637-650.
- Lani, R., Moghaddam, E., Haghani, A., Chang, L. Y., AbuBakar, S. and Zandi, K. (2014). Tick-borne viruses: a review from the perspective of therapeutic approaches. Ticks Tick Borne Dis 5(5): 457-465.
- Lazear, H. M. and Diamond, M. S. (2015). New insights into innate immune restriction of West Nile virus infection. Curr Opin Virol 11: 1-6.
- Liu, J., Elmore, J. M., Lin, Z. J. and Coaker, G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9(2): 137-146.
- Loo, Y. M., Fornek, J., Crochet, N., Bajwa, G., Perwitasari, O., Martinez-Sobrido, L., Akira, S., Gill, M. A., Garcia-Sastre, A., Katze, M. G. and Gale, M., Jr. (2008). Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82(1): 335-345.
- Loo, Y. M. and Gale, M., Jr. (2011). Immune signaling by RIG-I-like receptors. Immunity 34(5): 680-692.
- Izuogu, A. O., McNally, K. L., Harris, S. E., Youseff, B. H., Presloid, J. B., Burlak, C., Munshi-South, J., Best, S. M. and Taylor, R. T. (2017). Interferon signaling in Peromyscus leucopus confers a potent and specific restriction to vector-borne flaviviruses. PLoS One 12(6): e0179781.
- Robertson, S. J., Mitzel, D. N., Taylor, R. T., Best, S. M. and Bloom, M. E. (2009). Tick-borne flaviviruses: dissecting host immune responses and virus countermeasures. Immunol Res 43(1-3): 172-186.
- Santos, R. I., Hermance, M. E., Gelman, B. B. and Thangamani, S. (2016). Spinal cord ventral horns and lymphoid organ involvement in powassan virus infection in a mouse model. Viruses 8(8).
- Schoggins, J. W., Wilson, S. J., Panis, M., Murphy, M. Y., Jones, C. T., Bieniasz, P. and Rice, C. M. (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472(7344): 481-485.
- Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. and Schreiber, R. D. (1998). How cells respond to interferons. Annu Rev Biochem 67: 227-264.
- Suss, J. (2008). Tick-borne encephalitis in Europe and beyond--the epidemiological situation as of 2007. Euro Surveill 13(26).
- Taylor, R. T., Lubick, K. J., Robertson, S. J., Broughton, J. P., Bloom, M. E., Bresnahan, W. A. and Best, S. M. (2011). TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10(3): 185-196.
- Telford, S. R., 3rd, Armstrong, P. M., Katavolos, P., Foppa, I., Garcia, A. S., Wilson, M. L. and Spielman, A. (1997). A new tick-borne encephalitis-like virus infecting New England deer ticks, Ixodes dammini. Emerg Infect Dis 3(2): 165-170.
Article Information
Publication history
Accepted: Oct 29, 2017
Published: Dec 5, 2017
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Izuogu, A. O. and Taylor, R. T. (2017). Lentiviral Knockdown of Transcription Factor STAT1 in Peromyscus leucopus to Assess Its Role in the Restriction of Tick-borne Flaviviruses. Bio-protocol 7(23): e2643. DOI: 10.21769/BioProtoc.2643.
Category
Microbiology > Microbe-host interactions > Virus
Immunology > Host defense > Murine
Cell Biology > Cell signaling > Intracellular Signaling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link