- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Crosslinking Reactions and Substrate Incorporation Assays for The Identification of Transglutaminase-2 Protein Substrates
Published: Vol 10, Iss 12, Jun 20, 2020 DOI: 10.21769/BioProtoc.3657 Views: 2857
Reviewed by: Raquel Brandao HagaAleksei A TikhonovKossay Zaoui

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Site-specific Incorporation of Phosphoserine into Recombinant Proteins in Escherichia coli
Phillip Zhu [...] Richard B. Cooley
Nov 5, 2022 1303 Views
Measurement of Secreted Embryonic Alkaline Phosphatase
Meiyan Wang [...] Haifeng Ye
Feb 5, 2023 521 Views
A Modified Acyl-RAC Method of Isolating Retinal Palmitoyl Proteome for Subsequent Detection through LC-MS/MS
Sree I. Motipally [...] Saravanan Kolandaivelu
Apr 20, 2023 647 Views
Abstract
Transglutaminase (TG2) catalyzes protein crosslinking between glutamyl and lysyl residues. Catalytic activity occurs via a transamidation mechanism resulting in the formation of isopeptide bonds. Since TG2-mediated transamidation is of mechanistic importance for a number of biological processes, assays that enable rapid and efficient identification and characterization of candidate substrates are an important first-step to uncovering the function of crosslinked proteins. Herein we describe an optimized and flexible protocol for in vitro TG2 crosslink reactions and substrate incorporation assays. We have previously employed these techniques in the identification of the protein high mobility group box 1 (HMGB1) as a TG2 substrate. However, the protocol can be adapted for identification of any candidate transamidation substrate.
Keywords: TransglutaminaseBackground
Transglutaminase (TG2) is a member of the transglutaminase enzyme family that catalyzes calcium-dependent transamidation reactions. The transglutaminase enzyme family includes TG1-7 and FXIIIa, as well as band 4.2, a non-catalytically active member that plays a role in protein scaffolding (Satchwell et al., 2009; Gundemir et al., 2012). TG2 is unique to the family in that it is ubiquitously expressed and pleiotropic in function; in addition to protein crosslinking TG2 functions as a protein disulfide isomerase (Hasegawa et al., 2003; Mastroberardino et al., 2006), G-protein (Nakaoka et al., 1994; Baek et al., 1996; Vezza et al., 1999), and kinase (Mishra and Murphy, 2004; Mishra et al., 2006 and 2007).
Protein crosslinking occurs between the γ-carboxamide group of a peptide-bound glutamyl residue (a glutamine donor substrate) and the ϵ-amino group of a lysyl residue (lysine acceptor substrate), resulting in the formation of ϵ-(γ-glutamyl) lysine isopeptide bonds (Folk and Finlayson, 1977). A schematic of the crosslink reaction is shown in Figure 1.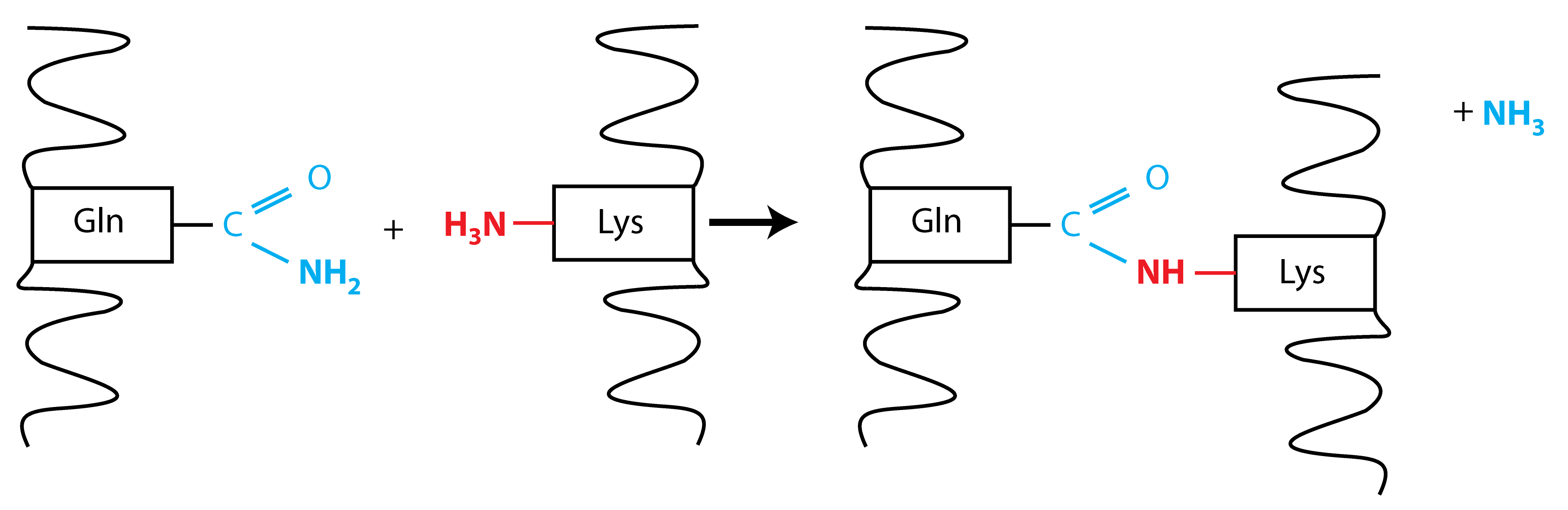
Figure 1. Schematic for the TG2 protein crosslink reaction. Protein crosslinking occurs by a transamidation reaction between the γ-carboxamide group of a peptide-bound glutamyl residue (shown in blue) and the ϵ-amino group of a lysyl residue (shown in red). Glutamine loses its amino group during the reaction, resulting in the formation of an ϵ-(γ-glutamyl) lysine isopeptide bond.
Although Tris-based buffers have been traditionally used for in vitro TG2 crosslink reactions, we found these buffers to be problematic for certain substrates, particularly those containing lysine donors only. Since Tris is a primary amine, it can potentially function as an acyl acceptor substrate in the transamidation reaction, competing with peptide-bound lysyl residues on protein substrates. To circumvent issues with Tris-based buffers, we instead used MOPS based buffer system for crosslink reactions. Since MOPSs is a tertiary amine, this avoids any potential issues with Tris- inhibition of the enzymatic reaction.
After confirmation that a protein is crosslinked by TG2, delineation of glutamyl vs. lysyl crosslink sites in the protein is an important next step to determine whether lysine and/or glutamine residues participate in the transamidation reaction.
The general scheme for in vitro crosslink reactions and substrate incorporation assays is described as follows:
1) Determine whether or not the protein of interest is a TG2 transamidation substrate.
Crosslink reactions between TG2 and purified recombinant forms of the putative substrate are performed in vitro and followed by Western blot analysis. This reaction can also be conducted in the context of endogenous proteins by using as substrates lysates from cell or tissue sources that contain the protein of interest instead of recombinant proteins. In either case, the appearance of a slower-migrating, size-shifted band by Western blot using antibodies against the putative substrate indicates a positive result. Regardless of whether purified recombinant proteins or endogenous sources from a cell or tissue lysate are included in the crosslink reaction, it is important to include the appropriate controls. The critical controls include reactions without TG2 enzyme, calcium, and/or using mutant TG2 enzyme lacking transamidation activity are for non-ambiguous data interpretation. Typical Western blot results for an in vitro crosslink reaction using purified recombinant HMGB1 are shown in Figure 2. Additionally, blocking peptide assays with the peptide antigen used to generate the antibody can be utilized to compete away the signals, confirming antibody specificity for TG2-modified substrates (Willis et al., 2018, Figure 3G).
2) Characterization of transamidation donor sites by substrate incorporation assays.
To determine whether the substrate in question undergoes crosslinking via glutamine or lysine donor residues or both, crosslink reactions between the purified recombinant proteins are supplemented with biotin-labeled glutamine- or lysine-donor probes. The crosslink reactions are then separated on SDS-PAGE and analyzed for biotin signal by Western blot with streptavidin-conjugated HRP. The utility of performing the substrate incorporation assay in this manner is that (in contrast to an ELISA-plate based format), additional qualitative information is gleaned about the size and relative abundance of probe-labeled reaction products on the gel. Additionally, specific bands of interest can be excised from the gel for protein identification or crosslink mapping by mass spectrometry (Willis et al., 2018).
Materials and Reagents
- 10 cm tissue culture plates (Corning, Catalog number: 430293 )
- 1.5 ml microcentrifuge tubes (USA Scientific, catalog number: 1615-5510 )
- 15 ml conical tubes (Thermo Scientific, catalog number: 339651 )
- Transglutaminase from guinea pig liver (Sigma-Aldrich, catalog number: T5398 )
- MOPS [3-(N-morpholino)propanesulfonic acid] (Sigma-Aldrich, catalog number: M1254 )
- Biotin-pentylamine (EZ-LinkTM Pentylamine-Biotin) (Thermo Scientific, catalog number: 21345 )
- Biotin-TVQQEL (a.k.a. A25 peptide) (Zedira, catalog number: B001 )
- Roche Complete protease inhibitor cocktail (Sigma, catalog number: 11697498001 )
- Dithiothreitol (DTT) (Sigma, catalog number: D9163 )
- PMSF (phenylmethanesulfonyl fluoride) (Sigma, catalog number: 78830 )
- Western blotting materials
- 4-12% polyacrylamide gels (Thermo Scientific catalog number: NW04120BOX )
- Mini gel tank (Thermo Scientific catalog number: A25977 )
- Nitrocellulose or PVDF membranes
- Electrophoretic transfer apparatus (GE Biosciences TE-22 )
- Tris [tris(hydroxymethyl)aminomethane] base (Fisher, catalog number: BP152-500 )
- HMGB1(high mobility group box protein 1; Genscript, catalog number: Z02803 )
- PBS (phosphate-buffered saline) (Thermo Scientific, catalog number: 10010049 )
- NaCl (Sigma-Aldrich, catalog number: S7653 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma, catalog number: E9884 )
- Sodium deoxycholate (Sigma, catalog number: D6750 )
- Sodium dodecyl sulfate (SDS) (Sigma, catalog number: L3771 )
- NP-40 (a.k.a. Igepal CA-630) (Sigma, catalog number: I8896 )
- Glycerol (Sigma, catalog number: G6279 )
- Streptavidin-Horse-radish peroxidase (Streptavidin-HRP)
- Protease inhibitor 25x stock solution (see Recipes)
- Dithiothreitol (DTT) 1 M stock solution (see Recipes)
- Crosslink reaction buffer (see Recipes)
- TG2 reconstitution buffer (see Recipes)
- Modified RIPA buffer (see Recipes)
- 6x SDS-loading buffer (see Recipes)
- Tris/SDS pH 6.8 (see Recipes)
Equipment
- Incubator or heating block (Fisher Scientific, catalog number: 88-860-022 )
- Electrophoretic transfer apparatus (GE Healthcare, catalog number: TE-22 )
- Refrigerated benchtop centrifuge (Eppendorf, model: 5424R )
- Gel box for running gels (mini gel tank) (Thermo Scientific, catalog number: A25977 )
- Platform shaker (VWR, catalog number: 10127-876 )
- Tissue culture incubator (Thermo Scientific, model: Steri-CycleTM i160)
- Western blot imaging equipment (film + developer machine, Chemi-Doc, Licor, etc.)
Procedure
- Preparation of cell lysates
- Remove cells from the tissue culture incubator and place on ice. For adherent cells growing in a 10 cm plate, discard tissue culture media and rinse the cell monolayer twice with 5 ml ice-cold PBS. For suspension cells, proceed to Step A3.
- While keeping the plate(s) on ice, add 1ml of ice-cold PBS, scrape and transfer the detached cells to pre-chilled microcentrifuge tubes on ice.
- Harvest the cells by centrifugation at 2,000 x g for 5 min at 4 °C. Discard the PBS supernatant.
- Lyse the cells by gently resuspending cell pellets in the appropriate amount of ice-cold modified RIPA lysis buffer (generally 200-500 μl for one 10 cm plate of 50-70% confluent cells) and incubate for 10 min on ice.
- Optional: If the cells were lysed in a buffer that releases chromatin from nuclei, sonication will be necessary to reduce sample viscosity. Keeping the cell lysates on ice, sonicate with 3-5 1-second pulses at 50% output.
- Centrifuge the lysates at 15,000 x g for 5 min at 4°C to pellet any insoluble cell debris.
- Transfer the supernatants to new microcentrifuge tubes pre-chilled on ice and assess the protein concentration by bicinchoninic acid (BCA) assay per the manufacturer’s instructions.
- In vitro TG2 crosslinking reactions
- Reconstitute the lyophilized TG2 enzyme at 4 U/ml (= 4 mU/µl) in TG2 reconstitution buffer. For a 1 unit vial of TG2, reconstitute with 250 µl of buffer for a final concentration of 4 U/ml. Place on ice while setting up the crosslink reactions.
- Prepare the in vitro crosslink reactions by adding the reagents in the order listed below to microcentrifuge tubes. The TG2 enzyme concentration, total reaction volume, and substrate concentrations should be optimized for each application. Some typical final concentrations for the reagents in reaction buffer are noted below:
- Reaction buffer with protease inhibitors
- Crosslink substrates: cell lysate (0.25 µg/µl) or purified recombinant proteins (10-20 ng/µl)
- TG2 enzyme (0.05-1.0 mU/ml)
- CaCl2 (2-5 mM)
- Incubate the reactions for 60 min at 37 °C.
- Quench the crosslink reactions by adding SDS-PAGE loading buffer to 1x concentration, followed by heating at 95 °C for 5 min on a heating block.
- Assess samples for protein crosslinking by SDS-PAGE followed by Western blotting.
When using in vitro crosslink reactions to test new potential TG2 substrates, positive and negative controls are also important. Using previously established crosslink substrates as a positive control will ensure the reactions are working as expected. Any commercially available protein with known TG2 substrate activity will work, with one caveat; the protein should have both glutamine and lysine crosslink sites. Otherwise a mixture of a protein with only glutamine donor sites and lysine acceptor sites would need to be included to ensure crosslinking occurs. Our lab typically uses recombinant Y-box 1 (YB-1) protein, which contains both glutamine and lysine crosslink sites and can be crosslinked to itself in vitro (Willis et al., 2013 and non-published data), or HMGB1, which contains only lysine acceptor sites but is crosslinked in vitro to Glutamine 636 of TG2 (Willis et al., 2018).
- Substrate Incorporation assays
For substrate incorporation assays, purified recombinant proteins are incubated with TG2 enzyme as described above, but in the presence of biotin-pentylamine (BP), a TG2 lysine donor substrate, or biotin-TVQQEL (A25 peptide), a TG2 glutamine donor substrate. Reaction mixtures are then size-fractionated on SDS-PAGE and analyzed for substrate incorporation with streptavidin -HRP by Western blot.
Reactions are set up in a similar manner to the TG2 crosslink reactions described above.- Reconstitute the lyophilized TG2 enzyme at 4 U/ml in TG2 reconstitution buffer. Place on ice while setting up the substrate incorporation assays.
- Set up the substrate incorporation reactions by adding the reagents to microcentrifuge tubes in the order they are listed below:
- Reaction buffer with protease inhibitors
- Purified recombinant protein crosslink substrate (10-20 ng/µl)
- BP or A25 peptide (2 mM)
- TG2 enzyme (1.0 mU/ml)
- CaCl2 (2-5 mM)
- Incubate for 60 min at 37 °C.
- Stop the reactions by adding concentrated SDS-loading buffer to a final 1x concentration and heat for 5 min at 95 °C on a heating block. The samples are ready to be separated by SDS-PAGE.
Data analysis
- TG2 crosslink reactions:
Run on SDS-PAGE and analyze by Western blot. TG2 substrate proteins will show a size-shift, often forming a slower migrating, higher molecular weight variant (or variants) near the top of the gel. This is often accompanied by a reduction in signal for the monomeric substrate (Willis et al., 2013 and 2018). Representative data for a crosslink reaction with recombinant HMGB1 as the substrate is shown in Figure 2.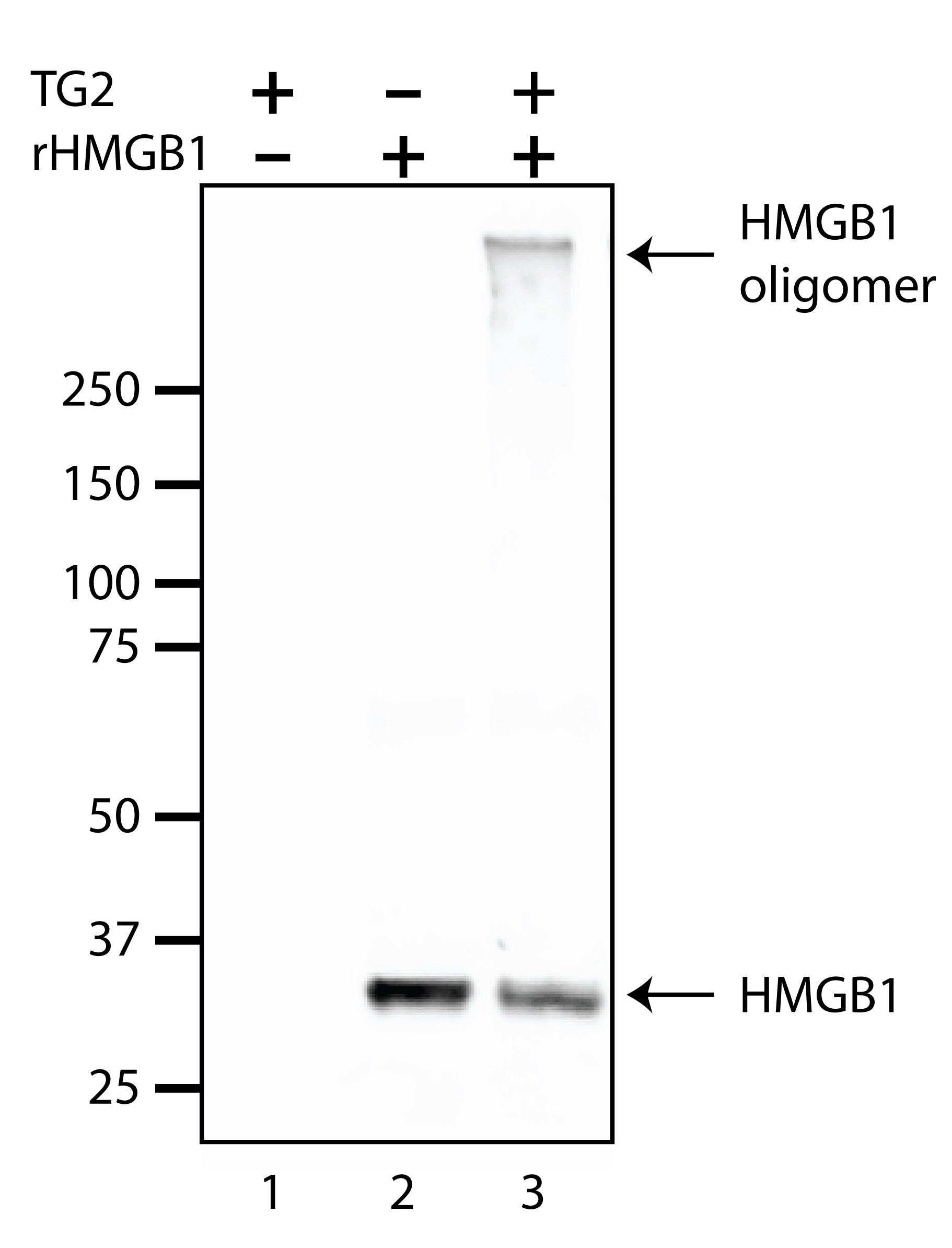
Figure 2. Representative Western blot results for an in vitro TG2 crosslink reaction. TG2 enzyme (0.2 mU/µl was incubated for 60 min at 37 °C in the presence of purified recombinant HMGB1 (rHMGB1) crosslink substrate (20 ng/µl) (Lane 3). TG2-only and recombinant HMGB1-only controls were loaded into lanes 1 and 2, respectively. Note that in lane 3, the ~29 kDa HMGB1 signal is diminished relative to lane 2, which coincides with the formation of the crosslinked HMGB1 oligomer near the top of the gel. Samples were separated on 4-12% polyacrylamide gels, transferred to nitrocellulose membranes and probed with HMGB1 antibody. - Substrate incorporation assays:
For the substrate incorporation assays, a reaction system using purified recombinant protein substrates facilitates straightforward analysis of the results by Western blot. Presence of a biotin-positive signal at the expected molecular weight for the substrate of interest indicates that biotinylated probes were incorporated into the substrate. Slower migrating/more retarded signals are indicative of the presence of multiple glutamyl and/or lysyl crosslink sites, since band migration is more retarded as the number of incorporated probe molecules increases (see Willis et al, 2018). It is also possible to excise proteins from the gel and map the specific crosslink sites by mass spectrometry, as noted in Willis et al., 2018.
The assay can also be performed in situ, where live cells are incubated with membrane-permeable biotinylated probes (Willis et al., 2018). After cell lysis, the biotin-tagged candidate substrate can be isolated with streptavidin beads; identification of the substrate by Western blot can be interpreted (in the context of the proper controls) as a positive result. Alternatively, proteins can be eluted from the streptavidin beads and subjected to mass spec analysis to determine the identity of the isolated proteins.
Notes
- A range of concentrations for cell lysate, TG2 enzyme, and/or biotinylated probes may be used. Final concentrations should be determined empirically. Note that higher concentrations of the biotinylated probes have been shown to function as inhibitors (Lorand et al., 1992), so care should be taken not to use excessive concentrations of labeled probes.
Depending on the abundance of transamidation substrate and other factors, a range of TG2 enzyme and protein substrate combinations should be tested with and without the biotin-labeled probes.
Although reconstituted TG2 will retain enzyme activity when stored at 4 °C for at least a couple weeks, it is best to reconstitute and use fresh enzyme the same day.
If the protein substrate of interest contains multiple transamidation donor sites, covalent addition of a number of probes will cause a size-shift on SDS-PAGE in proportion to the number of incorporated probe molecules. - Important controls for in vitro crosslinking reactions:
- No TG2 enzyme: reaction input (cell lysate or recombinant proteins) only. [For cell or tissue lysates with high levels of endogenous transamidation activity, this should be suppressed by adding 2 mM EGTA (a calcium-chelator) or 10 mM Iodoacetamide (an irreversible TG2 transamidation inhibitor)].
- TG2 enzyme only.
- No substrate: include reactions lacking the purified recombinant substrate, or cell or tissue lysates from sources where there is no expression or expression of the putative substrate is knocked out.
Recipes
- Protease inhibitor 25x stock solution
- Dissolve 1 tablet Roche Complete Protease inhibitor cocktail tablets in 2 ml distilled water
- Store small aliquots at -20 °C
- Dithiothreitol (DTT) 1 M stock solution
- Weigh 1.54 g of DTT and add to a 15 ml conical tube
- Add distilled water to 10 ml and dissolve completely
- Store small aliquots at -20 °C
- Crosslink reaction buffer
Note: Prepared fresh the day of the experiment; *add PMSF and DTT immediately prior to use.
50 mM MOPS, pH 7.5
0.2 mM PMSF*
1 mM DTT*
1x Protease inhibitor (Roche Complete Protease inhibitor cocktail) - TG2 reconstitution buffer
Note: Prepared fresh the day of the experiment; *add DTT immediately prior to use.
50 mM MOPS, pH 7.5
300 mM NaCl
0.5 mM EDTA
10 mM DTT* - Modified RIPA buffer
Note: Can be stored at 4°C after preparation; *add protease inhibitor, PMSF, and DTT immediately prior to use.
50 mM MOPS, pH7.5
150 mM NaCl
0.5% sodium deoxycholate
0.1% SDS
1% NP-40
0.2 mM PMSF*
1 mM DTT*
1x Protease inhibitor (Roche Complete Protease inhibitor cocktail) - 6x SDS-loading buffer (recipe for 20 ml)
Note: Prepare in advance and store at -20 °C in small aliquots.
14 ml Tris/SDS pH 6.8
6.0 ml glycerol
2 g SDS
1.86 g DTT - Tris/SDS pH 6.8 (for making 6x SDS loading buffer)
Note: Prepare in advance and store at room temperature.
50 ml 1 M Tris, pH 6.8
4 ml 10% SDS
46 ml dH2O
References
- Baek, K. J., Kwon, N. S., Lee, H. S., Kim, M. S., Muralidhar, P. and Im, M. J. (1996). Oxytocin receptor couples to the 80 kDa Gh alpha family protein in human myometrium. Biochem J 315 (Pt 3): 739-744.
- Folk, J. E. and Finlayson, J. S. (1977). The epsilon-(gamma-glutamyl)lysine crosslink and the catalytic role of transglutaminases. Adv Protein Chem 31: 1-133.
- Gundemir, S., Colak, G., Tucholski, J. and Johnson, G. V. (2012). Transglutaminase 2: a molecular Swiss army knife. Biochim Biophys Acta 1823(2): 406-419.
- Hasegawa, G., Suwa, M., Ichikawa, Y., Ohtsuka, T., Kumagai, S., Kikuchi, M., Sato, Y. and Saito, Y. (2003). A novel function of tissue-type transglutaminase: protein disulphide isomerase. Biochem J 373(Pt 3): 793-803.
- Lorand, L., Parameswaran, K. N., Velasco, P. T. and Murthy, S. N. (1992). Biotinylated peptides containing a factor XIIIa or a tissue transglutaminase-reactive glutaminyl residue that block protein cross-linking phenomena by becoming incorporated into amine donor sites. Bioconjug Chem 3(1): 37-41.
- Mastroberardino, P. G., Farrace, M. G., Viti, I., Pavone, F., Fimia, G. M., Melino, G., Rodolfo, C. and Piacentini, M. (2006). "Tissue" transglutaminase contributes to the formation of disulphide bridges in proteins of mitochondrial respiratory complexes. Biochim Biophys Acta 1757(9-10): 1357-1365.
- Mishra, S., Melino, G. and Murphy, L. J. (2007). Transglutaminase 2 kinase activity facilitates protein kinase A-induced phosphorylation of retinoblastoma protein. J Biol Chem 282(25): 18108-18115.
- Mishra, S. and Murphy, L. J. (2004). Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J Biol Chem 279(23): 23863-23868.
- Mishra, S., Saleh, A., Espino, P. S., Davie, J. R. and Murphy, L. J. (2006). Phosphorylation of histones by tissue transglutaminase. J Biol Chem 281(9): 5532-5538.
- Nakaoka, H., Perez, D. M., Baek, K. J., Das, T., Husain, A., Misono, K., Im, M. J. and Graham, R. M. (1994). Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science 264(5165): 1593-1596.
- Satchwell, T. J., Shoemark, D. K., Sessions, R. B. and Toye, A. M. (2009). Protein 4.2: a complex linker. Blood Cells Mol Dis 42(3): 201-210.
- Vezza, R., Habib, A. and FitzGerald, G. A. (1999). Differential signaling by the thromboxane receptor isoforms via the novel GTP-binding protein, Gh. J Biol Chem 274(18): 12774-12779.
- Willis, W. L., Wang, L., Wada, T. T., Gardner, M., Abdouni, O., Hampton, J., Valiente, G., Young, N., Ardoin, S., Agarwal, S., Freitas, M. A., Wu, L. C. and Jarjour, W. N. (2018). The proinflammatory protein HMGB1 is a substrate of transglutaminase-2 and forms high-molecular weight complexes with autoantigens. J Biol Chem 293(22): 8394-8409.
- Willis, W. L., Hariharan, S., David, J. J. and Strauch, A. R. (2013). Transglutaminase-2 mediates calcium-regulated crosslinking of the Y-box 1 (YB-1) translation-regulatory protein in TGFβ1-activated myofibroblasts. J Cell Biochem 114(12): 2753-2769.
Article Information
Publication history
Accepted: Apr 20, 2020
Published: Jun 20, 2020
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Willis, W. L., Foster, A., Henry, C., Wu, L. C. and Jarjour, W. (2020). In vitro Crosslinking Reactions and Substrate Incorporation Assays for The Identification of Transglutaminase-2 Protein Substrates. Bio-protocol 10(12): e3657. DOI: 10.21769/BioProtoc.3657.
- Willis, W. L., Wang, L., Wada, T. T., Gardner, M., Abdouni, O., Hampton, J., Valiente, G., Young, N., Ardoin, S., Agarwal, S., Freitas, M. A., Wu, L. C. and Jarjour, W. N. (2018). The proinflammatory protein HMGB1 is a substrate of transglutaminase-2 and forms high-molecular weight complexes with autoantigens. J Biol Chem 293(22): 8394-8409.
Category
Biochemistry > Protein > Posttranslational modification
Cell Biology > Cell-based analysis > Enzymatic assay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








