- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
HiSAT: A Novel Method for the Rapid Diagnosis of Allergy
Published: Vol 12, Iss 3, Feb 5, 2022 DOI: 10.21769/BioProtoc.4309 Views: 1426
Reviewed by: Alessandro DidonnaScott McCombSvetlana Kurilova

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Single Cell Migration Assay Using Human Breast Cancer MDA-MB-231 Cell Line
David M. Gau and Partha Roy
Apr 20, 2020 5605 Views
Spherical Invasion Assay: A Novel Method to Measure Invasion of Cancer Cells
Stephen D. Richbart [...] Piyali Dasgupta
Feb 20, 2022 3128 Views

Analysis of Mouse Brain Sections by Live-cell Time-lapse Confocal Microscopy
Tao Yang [...] Bing Ye
Apr 5, 2023 480 Views
Abstract
To identify causative substances for allergies to drugs or foods, the lymphocyte transformation test (LTT) is currently widely used as in vitro test, but its accuracy is not satisfactory. We have developed a novel method designated high-sensitivity allergy test (HiSAT) for determining allergy expression by measuring cell kinetics, using the chemotactic cells from non-allergic volunteers against a gradient field of cytokines released from immune cells when allergy develops. HiSAT requires a very small sample of 5 µL or less, and is applicable to three types of tests, depending on the situation in clinical practice: (i) diagnosis of the allergic expression, (ii) identification of the causative drug, and, in principle, (iii) pre-inspection.
Graphic abstract:
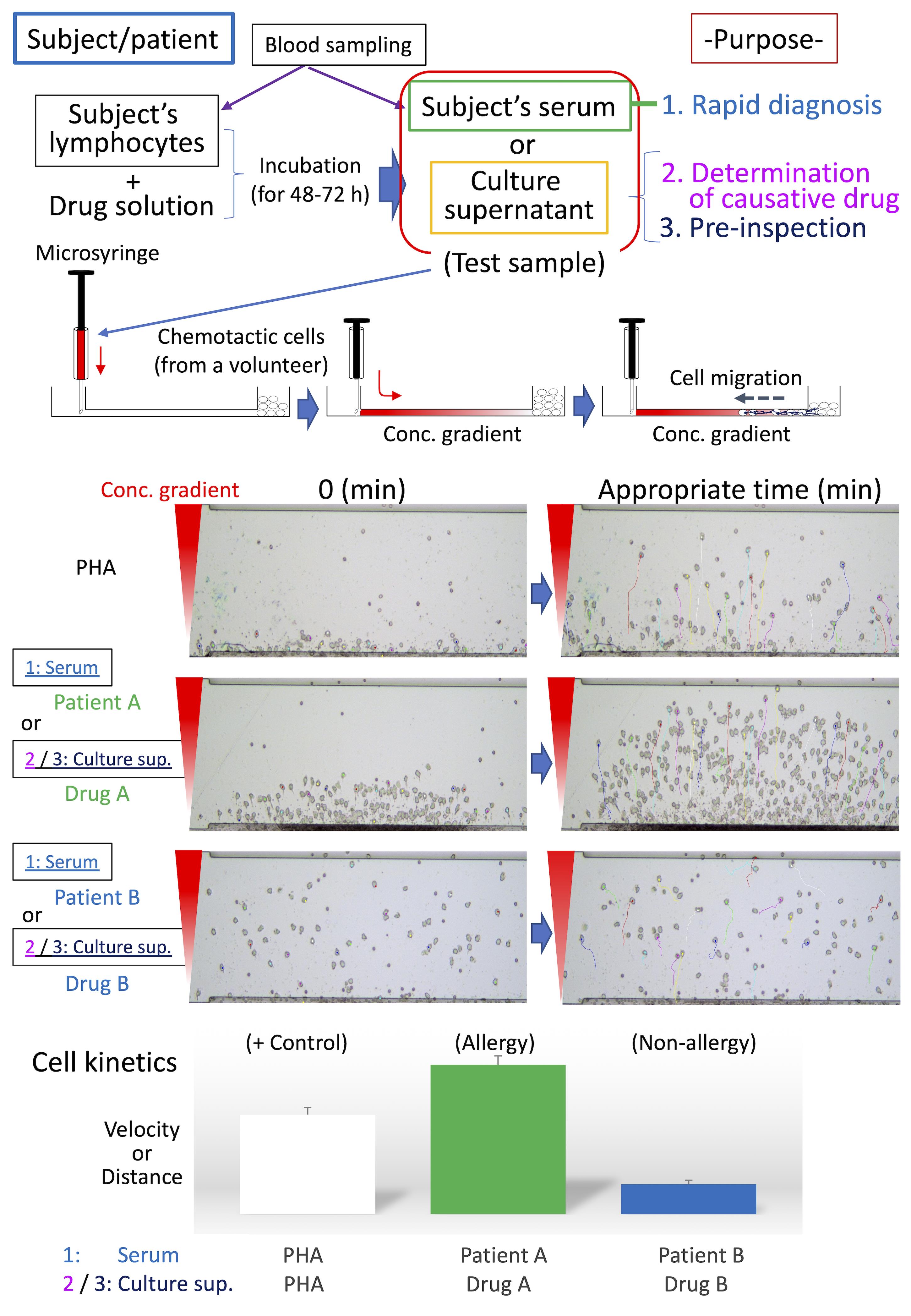
Schematic diagram of HiSAT. Serum from patients/subjects is used for rapid diagnosis in HiSAT. To identify the causative drug, the lymphocytes of interest are incubated with the candidate drug solution for 48 h to 72 h and then the culture supernatant is used in HiSAT. Before drug administration, it may possible to avoid the risk of allergies by performing pre-inspection, as well as the determination of the causative drug in HiSAT. A granulocyte-rich cell layer isolated from a non-allergic volunteer is used in HiSAT. Chemotactic cells migrate toward chemotactic factors in the test sample according to the concentration gradient. Cell kinetics (e.g., velocity or distance) are analyzed using sequential images of the test samples, and compared to the PHA-positive control.>
Background
Allergies, especially drug-induced allergy (DIA) or drug-induced hypersensitivity syndrome (DIHS), are unexpectedly caused by drugs used to treat the underlying disease, resulting in discontinuation of treatment or having to deal with allergic symptoms. Therefore, DIA is a heavy burden for both patients and medical personnel. Although it is strongly required in clinical practice, there is currently no rapid test to diagnose whether a patient's symptoms are allergic. On the other hand, once allergies are expressed in patients, it is necessary to identify the causative medicine. LTT is used as a general in vitro method for identifying the causative drug when DIA develops, and a specific antibody test such as RAST (Radioallergosorbent test) or MAST (Multiple allergosorbent test) is used against food-based or inhaled allergens. However, these tests have issues with accuracy or target limitation (Srinoulprasert, 2021). Furthermore, the conventional in vivo tests, such as the prick test, the scratch test, the intradermal test, and the patch test, lack accuracy in risk prediction. In addition, they might be burdensome on patients in positive cases, because of potentially serious allergic symptoms, such as anaphylactic shock. We have developed HiSAT, a novel allergy test, to diagnose allergy by analyzing cell kinetics of chemotactic cells from a non-allergic volunteer on the EZ-TAXIScan, using the subject's serum or the culture supernatant incubating the subject's lymphocytes and candidate drug (Shibaguchi et al., 2021). The advantages of HiSAT are as follows: (i) the amount of the sample is very small (5 µL or less is required), (ii) rapid diagnosis of about 1 hr or less is possible if using subject serum, (iii) the reactivity of each subject is directly confirmed in vitro, (iv) it is possible to test even new substances that have not been reported so far, (v) it does not depend on the expression mechanism (Warrington et al., 2018) because it targets the humoral components released when allergy develops, and (vi) in principle, it is possible to use for pre-inspection (Figure 1).
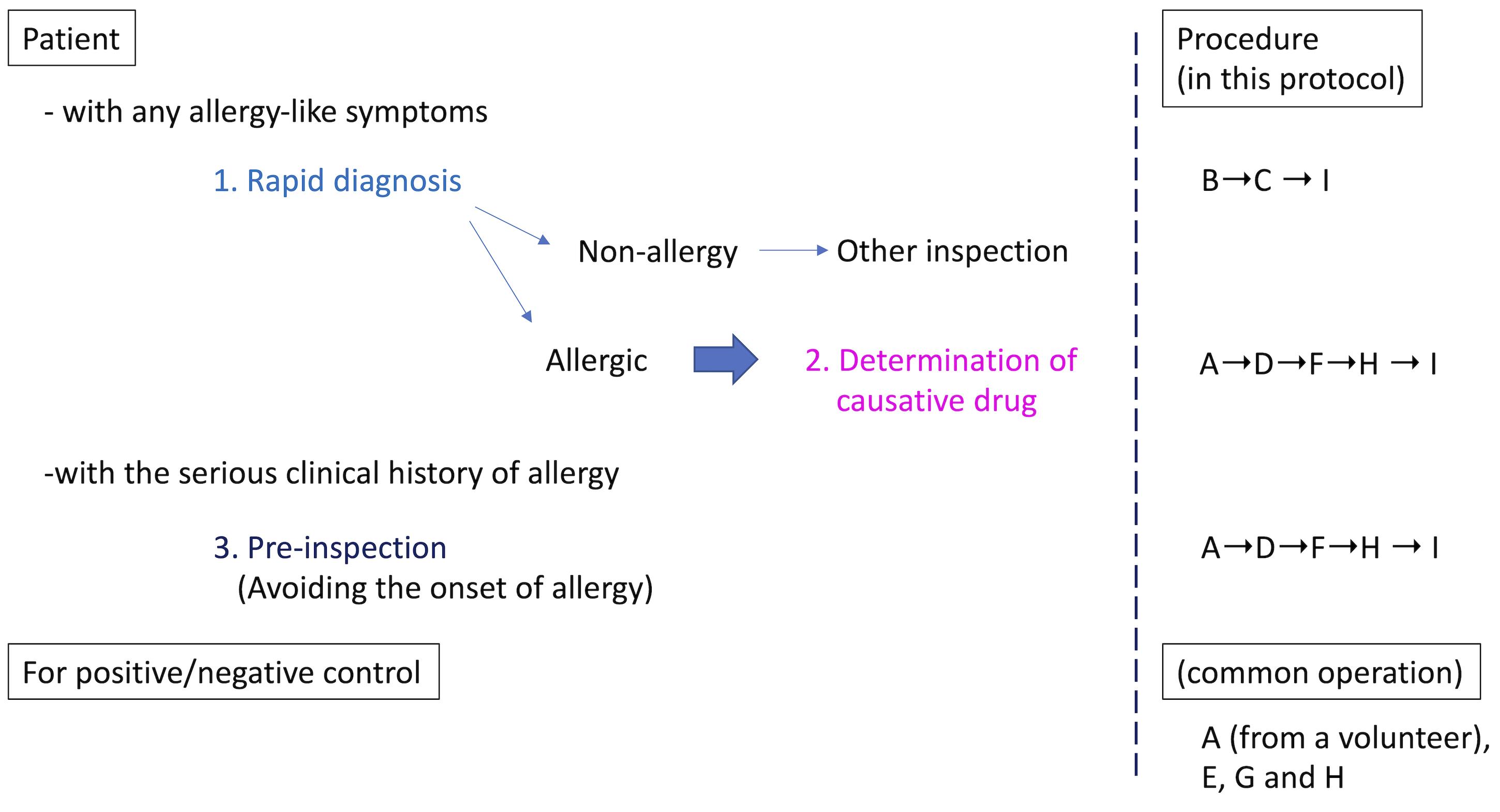
Figure 1. Schematic procedure in HiSAT. If there are any allergy-like symptoms in the patient, the rapid test using the patients' serum is useful. There is no such rapid test so far. Once the symptom is diagnosed as allergic, it is important to determine the causative drug. On the other hand, if a patient has a serious clinical history of allergy, pre-inspection might be useful, to avoid the dangerous drug before use. In all cases, the positive and negative controls have to be prepared in advance.
Materials and Reagents
1-mL syringe (TERMO Co. Ltd., catalog number: SS-01T)
1.5 mL microtube (Funakoshi Co. Ltd., catalog number: 1210-00)
15 mL Falcon tube (Corning, catalog number: 352096)
Pipette tip (P20, P100, P1000: Greiner, catalog numbers: 771351, 770320, 770340)
Pipette (GILSON, M&S Instruments Inc., catalog numbers: F144802, F123601, F123602)
Pasteur pipette (Funakoshi Co. Ltd., cattalog number: 63B92)
10-µL hamilton microsyringe (GL Science, catalog number: 4015-63015)
4 µm thickness of tip for EZ-TAXIScan (GE Healthcare Bioscience Co. Ltd., catalog number: EZT-01-4)
24-well plates (BD Falcon, Corning, catalog number: 353047)
Phosphate buffered saline (PBS, pH 7.4; Thermo Fisher Scientific, Gibco, catalog number: 158127)
RPMI 1640 (pH 7.4, Sigma-Aldrich, catalog number: R8758)
Dalbecco Modified Eagle's Medium (DMEM, pH 7.4, Sigma-Aldrich, catalog number: D6429)
Horse serum (Thermo Fisher Scientific, Gibco, catalog number: 26050070)
Fetal bovine serum (FBS; Fujifilm Wako Chemicals Co.,Ltd., catalog number: FB-1345)
Penicillin (10,000 units) and streptomycin (10 mg) (Sigma-Aldrich, catalog number: P4333)
Venoject II (Terumo Coop., catalog number: VP-AS109K)
Phytohemagglutinin (PHA; Fujifilm Wako Chemicals Co.,Ltd., catalog number: 161-15251)
Lymphoprep (Cosmo Bio Co.Ltd., catalog number: 331312)
Lymphocyte-poly Cell Separation Media (Cosmo Bio Co. Ltd., catalog number: CL5070)
Dimethyl sulfoxide (DMSO; Sigma-Aldrich, catalog number: D2650)
Mosapride citrate hydrate (Sumitomo Dainipon Pharma Co. Ltd., KEGG drug: D00724)
Sulpiride (Nichi-iko Pharmaceutical Co. Ltd., KEGG drug: D01226)
Rabeprazole sodium (Eisai Co, Ltd., KEGG drug: D00724)
Equipment
Centrifuge (TOMY Co. Ltd., catalog number: NIX-521)
Incubator (37°C, 5% CO2) (Thermo Fisher Scientific, catalog number: 4130TS)
Refrigerator (NIHON Freezer Co. Ltd., catalog number: KGT-4056HC)
Freezer (NIHON Freezer Co. Ltd., catalog number: VT-208)
Inverted microscope (Olympus Co. Ltd., catalog number: CKX-53)
EZ-TAXIScan system (GE healthcare Bioscience Co. Ltd., catalog number: MIC-1001)
Software
ImageJ software: Image Processing and Analysis in Java, "https://imagej.nih.gov/index.html", National Institutes of Health, Bethesda, MD, USA
Chemotaxis and Migration Tool, "https://ibidi.com/chemotaxis-analysis/171-chemotaxis-and-migration-tool.html", ibidi GmbH, Gräfelfing, Bayern, DE
GraphPad Prism software: "https://www.graphpad.com/scientific-software/prism/", GraphPad software, La Jolla, CA, USA
Procedure
PBMC separation
Peripheral blood mononuclear cells (PBMC) are extracted from whole-blood samples of the subjects, using density-gradient centrifugation with Lymphoprep solution (22°C).
Add Lymphoprep solution to 15 mL tube.
Dilute blood with an equal volume of PBS.
Layer blood on top of Lymphoprep, being careful to minimize mixing of blood with Lymphoprep.
Centrifuge at 800 × g for 20 min at room temperature, using a swing rotor.
Transfer mononuclear cell layer under plasma layer using a Pasteur pipette, without disturbing tha plasma-Lymphoprep interface to new tube.
After washing with 10 mL of ice-cold PBS twice, the cells are resuspended at a concentration of 1.25 × 106 cells/mL in RPMI 1640 medium (37°C) supplemented with horse serum (5%), penicillin (100 U/mL) and streptomycin (100 µg/mL).
Chemotactic cells
The granulocyte-rich cell layer (chemotactic cells) from the blood samples of the volunteer without allergic symptoms are separated using Lymphocyte-poly cell separation media (22°C).
Carefully layer 5 mL of whole blood on top of an equal volume of Lypmpholyte-poly in a 15 mL tube.
Centrifuge at 500 × g for 30 min in a swing rotor at room temperature.
Harvest the polymorphonuclear cell (chemotactic cell) band (lower band) and transfer new tube using a Pasteur pipette.
Wash the cells with PBS by centrifugation at 400 × g for 10 min at room temperature.
Suspend the chemotactic cells in DMEM medium (22°C) at a concentration of 1 × 107 cells/mL.
Serum separation
Separate the subject's serum from whole-blood samples using a Venoject II tube.
After blood sampling, leave tubes for over 1 h at room temperature, followed by centrifugation for 10 min at 350 × g at room temperature.
Transfer the serum on the top into the falcon tube and, if necessary, store at -20°C until further use.
Drug solution
The drug solutions contain the test drug (Mosapride, Sulpiride or Rabeprazole) at a final concentration of one-half of the Cmax (15, 75, or 200 ng/mL, respectively) in PBS, except for Sulpiride (in DMSO).
PHA solution
Prepare the PHA solution by dissolving PHA in PBS at a final concentration of 1 µg/mL.
Preparation of antigen solution
Prepare the sntigen solutions by mixing the drug solution (50 µL) and the subject's serum (50 µL).
Control solutions
Instead of the drug solution, 50 µL of PBS or PHA solution are used as the negative and positive controls, respectively.
Preparation of test sample (culture supernatant)
In 24-well plates, stimulate the 1.9 mL (2.4 × 106 cells) of PBMC by addition of 100 µL of antigen solution or control solution.
Culture PBMC for 48-72 h at 37°C in a CO2 incubator.
Transfer the culture supernatants to 15 mL tubes and centrifuge at 400 × g for 5 min at room temperature, collect them in new tubes as test samples, and store them at -20°C until further use.
Real-time monitoring of cell migration
Record cell migration against serum or culture supernatants over time using EZ-TAXScan (Kanegasaki et al., 2003).
Mount the glass plate on the bottom base of the holder, and fill the house with 4 mL of RPMI medium (37°C).
Mount the silicon substrate and wafer clamp in the house on the glass plate and press them down with the mechanical clamp.
Make the contacts between the lower surface of the etched substrate to the glass plate and the upper surface to the highly polished stainless steel clamp water-tight with mechanical pressing, due to the optical flatness of the surfaces.
Remove the air bubbles trapped in the observation area or the compartment of each channel when assembled, if any, by pushing and pulling the medium with a 1 mL syringe.
After assembling and setting the observation unit in the appropriate position on EZ-TAXIScan, load the chemotactic cells into the lower (front) wells at 5-10 µL (5-10 × 104 cells)/well (Figure 1A).
Shift the chemotactic cells near the observation area by aspiration with a 1 mL syringe.
Subsequently, add the serum or culture supernatant (1.5 µL/well), depending on the purpose, to the upper (back) wells with a 10 µL-microsyringe, thereby producing the gradient (gradient length, 260 µm).
The observation stage is 4 µm in-depth for human granulocyte-rich cells.
Record the images of chemotactic cells in the observation area at 2-min intervals over the 240-min experimental period using a computer connected to the microscope.
The chemotactic cells migrate toward the upper (back) well from the lower (front) well depending on the reactivity of humoral chemotactic factors (Figure 2).
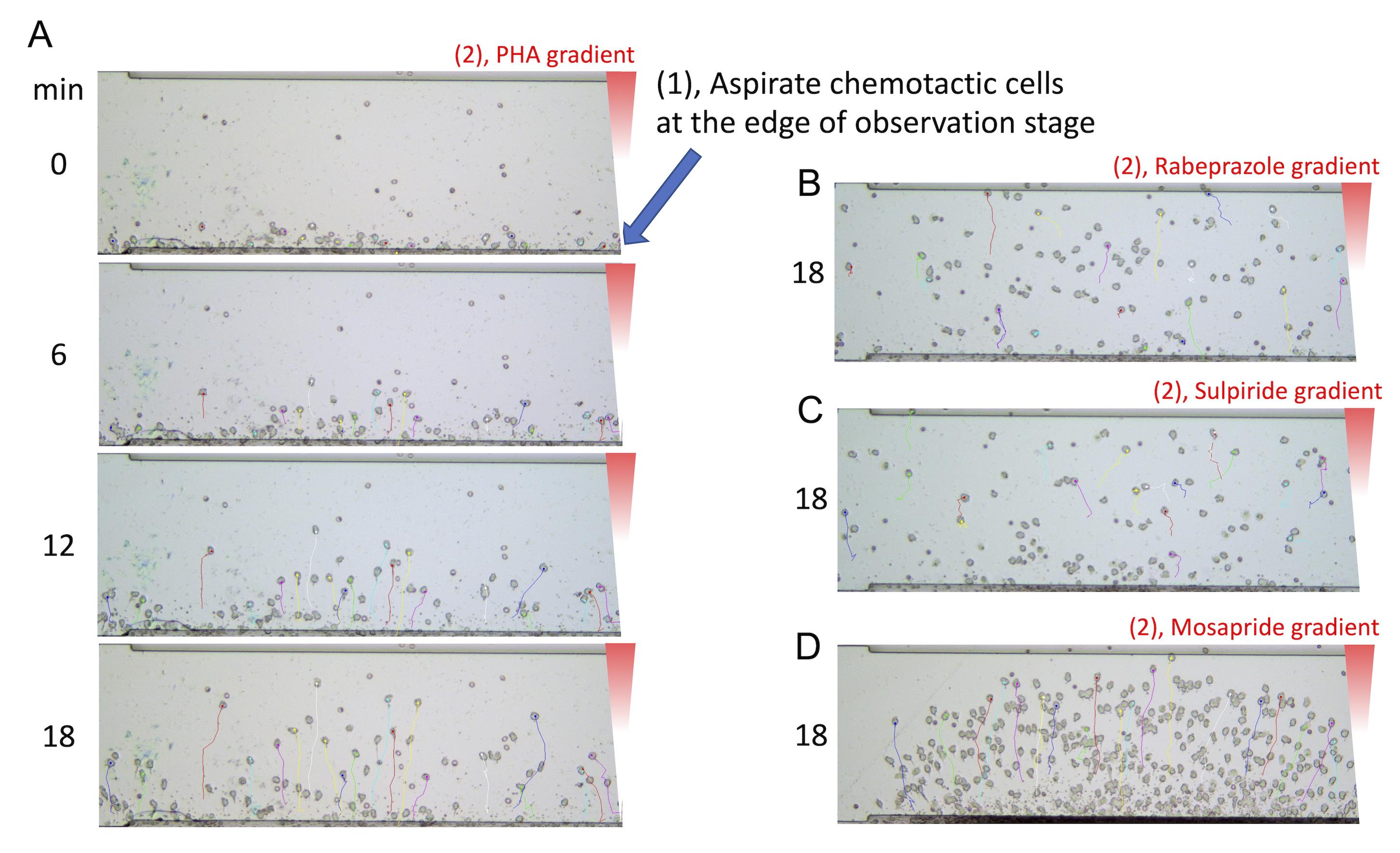
Figure 2. Typical images of cell migration in HiSAT. The chemotactic cells separated from the non-allergic volunteer are aligned at the edge of the observation stage by aspiration with a syringe (A). By reacting the subject's lymphocytes with the drug solution, the cells migrate according to the concentration gradient of the culture supernatant containing the chemotactic factors (A-D). Similar images can be obtained by using serum instead of culture supernatant.
Data analysis
Calculation of cell kinetics
Determine the cell migration velocity and the distance traveled by analyzing 10-16 migrating cells selected randomly, using the ImageJ software plugin "Manual Tracking"(Figure 1). Select the appropriate time range for analyzing cell kinetics in HiSAT (0 to 18 min, 2-min intervals in Figure 1). The "Chemotaxis and Migration Tool" provides the numerical data of velocity and distance based on the experimental data. Exported ASCII (mandatory format) tables from "manual tracking" are directly imported into the software tool.
Statistical analysis
Numerical data are presented as the mean± standard error. Significant differences between different treatment groups are analyzed using one-way analysis of variance, followed by Bonferroni's multiple comparison tests or Student's t-test using GraphPad Prism software. P-value <0.05 was considered statistically significant.
Acknowledgments
This study was supported in part by the fund [grant number 27-8-3] from the Kakihara Science and Technology Research Foundation and by the funds (grant numbers 116005, 146003 and 187003) from the Central Research Institute of Fukuoka University. This study was also supported in part by JSPS KAKENHI (grant number 17K15534).
Competing interests
There are no conflicts of interest to declare.
Ethics
The patients suspected of having cryptogenic allergy-like symptoms were hospitalized at Fukuoka University Hospital (Fukuoka. Fukuoka, Japan) from April in 2011 to December in 2017, and the volunteers were enrolled in this study. The volunteers were the hospitalized patients without allergic symptoms in Fukuoka University Hospital and signed the consent form to use wasted blood of regular blood sampling. This study was approved by the Institutional Review Board of Fukuoka University Hospital (approval no. IRB11-4-16). Additionally, written informed consent was obtained from each patient before enrolment.
References
- Kanegasaki, S., Nomura, Y., Nitta, N., Akiyama, S., Tamatani, T., Goshoh, Y., Yoshida, T., Sato, T. and Kikuchi, Y. (2003). A novel optical assay system for the quantitative measurement of chemotaxis. J Immunol Methods 282(1-2): 1-11.
- Shibaguchi, H., Yasutaka, Y. and Futagami, K. (2021). Novel method to analyze cell kinetics for the rapid diagnosis and determination of the causative agent in allergy. PLoS One 16(2): e0246125.
- Srinoulprasert, Y. (2021). Lymphocyte transformation test and cytokine detection assays: Determination of read out parameters for delayed-type drug hypersensitivity reactions. J Immunol Methods 496: 113098.
- Warrington, R., Silviu-Dan, F. and Wong, T. (2018). Drug allergy. Allergy Asthma Clin Immunol 14(Suppl 2): 60.
Article Information
Publication history
Accepted: Nov 30, 2021
Published: Feb 5, 2022
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shibaguchi, H. and Yasutaka, Y. (2022). HiSAT: A Novel Method for the Rapid Diagnosis of Allergy. Bio-protocol 12(3): e4309. DOI: 10.21769/BioProtoc.4309.
Category
Immunology > Inflammatory disorder > Allergy
Medicine
Cell Biology > Cell movement > Cell migration
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







