- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Colocalization Assay with Fluorescent-tagged ATG8 Using a Nicotiana benthamiana-based Transient System
Published: Vol 12, Iss 16, Aug 20, 2022 DOI: 10.21769/BioProtoc.4486 Views: 1635
Reviewed by: Wenrong HeYuan WangYe XuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Protocol for Mitotic Metaphase Chromosome Count Using Shoot Meristematic Tissues of Mulberry Tree Species
Raju Mondal [...] K. Vijayan
Sep 5, 2023 387 Views

Synthetic Promoter Screening Using Poplar Mesophyll Protoplast Transformation
Yongil Yang [...] C. Neal Stewart Jr.
Apr 20, 2023 573 Views
An In-depth Guide to the Ultrastructural Expansion Microscopy (U-ExM) of Chlamydomonas reinhardtii
Nikolai Klena [...] Virginie Hamel
Sep 5, 2023 1042 Views
Abstract
Autophagy is an evolutionarily conserved intracellular degradation process. During autophagy, a set of autophagy-related (ATG) proteins orchestrate the formation of double-bound membrane vesicles called autophagosomes to engulf cytoplasmic material and deliver it to the vacuole for breakdown. Among ATG proteins, the ATG8 is the only one decorating mature autophagosomes and therefore is regarded as a bona fide autophagic marker; colocalization assays with ATG8 are wildly used as a reliable method to identify the components of autophagy machinery or autophagic substrates. Here, we describe a colocalization assay with fluorescent-tagged ATG8 using a tobacco (Nicotiana benthamiana)-based transient expression system.
Keywords: AutophagyBackground
As a highly conserved catabolic process, autophagy is sophisticatedly employed by eukaryotic cells to remove dysfunctional proteins and unwanted or damaged organelles via the vacuole in yeast and plants or the lysosome in animals. During autophagy, a set of ATG proteins, commonly referred to as core autophagic machinery, are recruited to direct the formation of autophagosomes (Marshall and Vierstra, 2018). Among them, ubiquitin-like protein ATG8 (or its mammalian homolog, LC3) is first lipidated through a ubiquitination-like enzymatic cascade and then coats expanding autophagic vesicles to promote the maturation of autophagosomes. Moreover, the lipidated ATG8 on the inner membrane of autophagosomes can serve as a docking platform for autophagy receptors to recruit specific cargoes during selective autophagy. When autophagosomes fuse with the vacuole or lysosome, the lipidated ATG8 lining the outer membrane is released by ATG4 and recycled, whereas the lipidated ATG8 on the inner membrane is degraded together with cargoes into the vacuole. Therefore, ATG8 is regarded as a bona fide marker for autophagosomes, and colocalization assays with ATG8 are wildly used as a reliable method for identification of ATG components or autophagic cargoes (Marshall and Vierstra, 2018).
Colocalization assays with fluorescent-tagged ATG8 have been carried out stably with transgenic plants or transiently with a protoplast expression system (Suttangkakul et al., 2011; Zhuang et al., 2013). Here, we have developed an alternative colocalization assay based on a tobacco (Nicotiana benthamiana) transient expression system. With this system, we have successfully detected the colocalization between ATG8 and multiple proteins of interest (POIs; Luo et al., 2021).
This protocol includes four major steps: 1) Vector construction; 2) Agrobacterium-mediated transient expression; 3) Concanamycin A (ConA) infiltration; 4) Confocal fluorescence microscopy imaging.
Materials and Reagents
1.5 mL microcentrifuge tubes (Biosharp, catalog number: BS-15-M)
50 mL mini bioreactor tubes (NEST, catalog number: 602052)
9 cm Petri dish (Sangon Biotech, catalog number: F611001)
1 mL syringe
Microscope slides, 76 × 26 mm (Sangon Biotech, catalog number: F518101)
Cover slides, 24 × 60 mm (Sangon Biotech, catalog number: F518118)
Marker pen
Single-edge razor blade
Aluminum foil
4-week-old Nicotiana benthamiana seedlings
7-day-old Arabidopsis seedlings
Agrobacterium tumefaciens strain GV3101
E. coli DH5α competent cells (Home-made)
LB (Sangon Biotech, catalog number: A507002)
Sucrose (Sangon Biotech, catalog number: A610498)
MgSO4 (Sangon Biotech, catalog number: A601988)
Agar (Sangon Biotech, catalog number: A505255)
MES (Sangon Biotech, catalog number: A610341)
0.5 M EDTA solution, pH 8.0 (Sangon Biotech, catalog number: B540625)
Acetosyringone (Sangon Biotech, catalog number: A601111)
Dimethyl sulfoxide (DMSO) (Sangon Biotech, catalog number: A100231)
MS basal salt mixture (Caisson, catalog number: 05210003)
Concanamycin A (Adipogen, catalog number: BVT-0237-C100)
Plant RNAout kit (Tiandz Inc., catalog number: 160906-50)
HiScript® II Q RT SuperMix for qPCR Kit (Vazyme Biotech Co., Ltd, catalog number: R222-01)
HiPure Gel DNA Mini Kit (Magen, catalog number: D2111-03)
ClonExpress® II One-Step Cloning Kit (Vazyme Biotech Co., Ltd, catalog number: C112-01)
pEGAD vector (NCBI accession No. AF218816)
FastDigest EcoRI-HF (NEB, catalog number: R3101M)
FastDigest BamHI-HF (NEB, catalog number: R3136M)
Antibiotics (All purchased from Sangon Biotech; Kanamycin sulfate, catalog number: A506636 Rifampicin, catalog number: A600812; Gentamycin sulfate, catalog number: A506614)
Liquid nitrogen
Bacterial suspension buffer (see Recipes)
ConA stock solution (0.5 mM) (see Recipes)
MS-C liquid medium (see Recipes)
Equipment
Centrifuge (Eppendorf, model: 5804 R)
Water bath
Eppendorf Research® plus Pipette (0.5–10 μL pipette, catalog number: 3120000020; 10–100 μL pipette, catalog number: 3120000046; 100–1,000 μL pipette, catalog number: 3120000062)
Nanodrop spectrophotometer (NanoDrop, model: ND-1000)
Incubator
Flow hood
Plant growth chamber (Ningbo Jiangnan Instrument Factory, model: RXZ-500C)
pH meter (Sartorius, model: PB-10)
Thermocycler (Bio-Rad, model: S1000)
Autoclave (HIRAYAMA, model: HVE-50)
Vortexer (Kylin-Bell, model: VORTEX-5)
Procedure
Vector construction
PCR amplification of the Arabidopsis AtATG8a (AT4G21980) gene: Isolate total RNA from 7-day-old seedlings using the Plant RNAout kit (Tiandz Inc.,160906-50), and then convert 1 μg of total RNA to cDNA using the HiScript® II Q RT SuperMix for qPCR Kit (Vazyme Biotech Co., Ltd, R222-01) with oligo(dT)20. Amplify the AtATG8a gene using PCR with the forward primer 5’-CTGCGGCAGCGGCCGAATTCATGGCTAAGAGTTCCTTCAA-3’ and the reverse primer 5’-ATCTAGATCCGGTGGATCCATCCAAAAGTGTTCTCTCCAC-3’ (40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s).
Note: PCR primers should be designed to contain at least 20 bp overlapping sequences (underlined) to a linearized cloning pEGAD vector (see Figure 1A).
Cloning the AtATG8a gene into the pEGAD vector: Linearize the pEGAD vector (NCBI accession No. AF218816) with the restriction enzymes EcoRI and BamHI at 37 °C for 4 h, and then purify with the HiPure Gel DNA Mini Kit (Magen, D2111-03). Assemble the linearized pEGAD vector and the amplified AtATG8a fragment using the ClonExpress® II One-Step Cloning Kit following the manufacturer’s instructions (Vazyme Biotech Co., C112-01). Construct the vector containing protein of interest (POI, pEGAD-POI-mCherry) using a similar method (Figure 1C).
Note: For each POI, two constructs should be designed, with one containing a fluorescent protein at its N−terminus, and the other at the C−terminus. Some general guidelines for creating fluorescent fused protein in plants can be found in Tanz et al. (2013).
E. coli transformation: Mix the 10 μL of the above reaction solution with 100 μL E. coli DH5α competent cells and incubate on ice for 30 min. Heat the cells in a 37 °C water bath for 90 s and incubate on ice for 1–2 min. Add 700 μL of LB to the cells and shake at 37 °C, 200 rpm for 1 h for recovery. Centrifuge at 5,000 rpm for 3 min, then discard 600 μL of supernatant and resuspend the pellet in the remaining supernatant. Spread on an LB plate containing 50 µg/mL of kanamycin and incubate for 12–14 h at 37 °C. Perform colony PCR to identify positive clones and verify by Sanger sequencing with primer: 5’-AATCATCGCAAGACCGGCAACAGGAT-3’.
Agrobacterium transformation: Mix 1 µg of verified plasmid with 200 μL GV3101 competent cells and incubate on ice for 30 min. Freeze the cells with liquid nitrogen for 1–2 min and then thaw by incubating in a 37 °C water bath for 2 min. Add 1 mL of LB to the cells and shake at 28 °C, 200 rpm for 4 h for recovery. Centrifuge at 7,000 rpm for 1 min, discard 1 mL of supernatant, and resuspend the pellet in the remaining supernatant. Spread the cells on LB plates containing appropriate antibiotics and incubate at 28 °C for 2 days for colony formation.
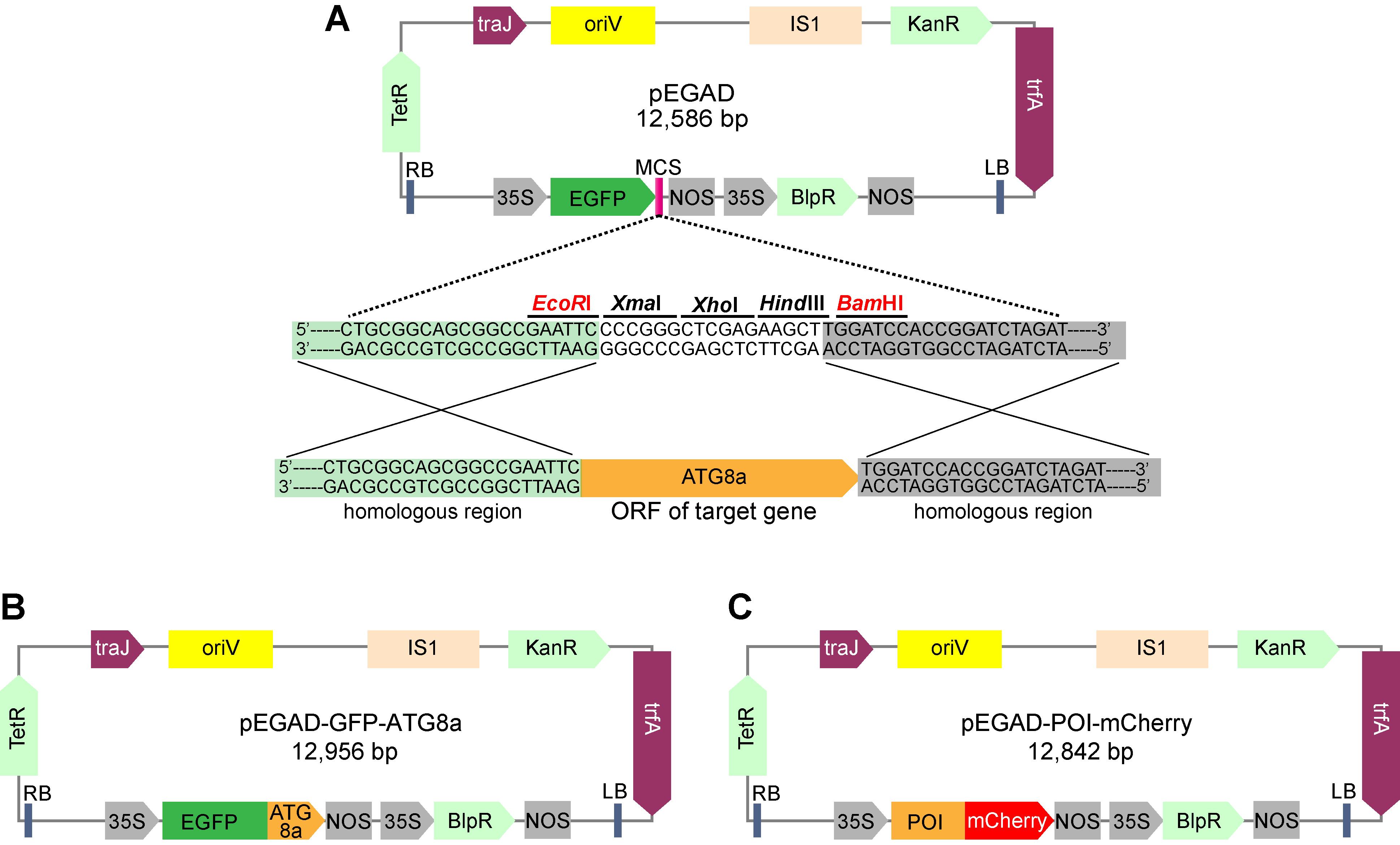
Figure 1. Schematic diagram of the vector constructs used in ATG8 colocalization assay in N. benthamiana leaves. (A) Construction of pEGAD-GFP-ATG8a vector using one-step cloning method. (B) and (C) are the schematic diagrams of pEGAD-GFP-ATG8a and pEGAD-POI-mCherry vectors, respectively. Promoters, terminators, and open reading frames are indicated in the maps. 35S: Cauliflower mosaic virus 35S promoter; BlpR: Basta resistance gene; IS1: insertion sequence; KanR: Kanamycin resistance gene; LB: left border; NOS: NOS terminator; oriV: origin of replication; POI, protein of interest. RB: right border; TetR: repressor of the tetracycline resistance element; trfA: replication initiator protein; traJ: Positive regulator of the F plasmid transfer (tra) operon.
Agrobacterium-mediated transient gene expression in tobacco leaf
Pick up a single A. tumefaciens colony for each construct and inoculate into 15 mL of LB liquid medium containing appropriate antibiotics (50 μg/mL of kanamycin, 50 μg/mL of rifampicin, 10 μg/mL of gentamycin) in a 50 mL mini bioreactor tube. Shake at 200 rpm, 28 °C for 16–24 h until OD600 = 0.8–1.5.
Harvest the cells by centrifugation at 6,000 rpm, 4 °C, for 10 min. Resuspend the cells with fresh suspension buffer (see Recipes) to a final concentration of OD600 = 1.2 and keep at 25 °C for more than 4 h before infiltration (Figure 2A).
Note: As a common practice, acetosyringone is added into bacterial cultures to activate Agrobacterium virulence. The optimal acetosyringone (150 µM) treatment usually requires at least 4 h.
Mix equal volumes of the cells containing the pEGAD-GFP-ATG8a and pEGAD-POI-mCherry or negative control pEGAD-mCherry constructs, and infiltrate on the abaxial side of the tobacco leaf using a 1 mL needleless syringe. Each half of the leaf is infiltrated with about 100 μL of bacterial mixture (Figure 2B), and the infiltrated areas are labeled with a marker pen (Figure 2C).
Note: Infiltrate tobacco leaf carefully to avoid damaging leaf.
Keep the infiltrated tobacco seedlings in a growth chamber at 28 °C under 16/8 h light/dark for 24–36 h.
Concanamycin A (ConA) infiltration
Excise a small leaf disc close to the site of infiltration and examine the expression of GFP or mCherry-tagged proteins in a fluorescence microscope.
Infiltrate diluted ConA solution (1 μM) with 1 mL needleless syringe over the marked leaf areas (Figure 2D). Cut the leaves when ConA solution is almost absorbed (usually takes about 15 min, Figure 2E).
Note: ConA is a specific inhibitor of vacuolar-type ATPases and is often used to stabilize the autophagic bodies (Dröse et al., 1993; Dettmer et al., 2006). The diluted ConA solution must be prepared freshly before use by adding 998 μL fresh suspension buffer to 2 μL ConA stock solution (0.5 mM). Moreover, it is important to use a different injection site for ConA infiltration.
Remove the midvein of detached leaves and place into Petri dishes containing two layers of filter paper pre-wetted with MS liquid medium without sugar (MS-C medium, see Recipes) (Figure 2F). Cover the leaves with one more layer of pre-wetted filter paper and gently press against the leaves to keep them wet (Figure 2G).
Note: Carbon starvation is a common approach for autophagy induction. To do so, MS-C liquid medium is used here, and ConA-infiltrated leaves are cut and placed in the dark. Moreover, add about 1.5 mL of MS-C medium to wet filter paper. Too much water will cause cell death in the leaves.
Cover the Petri dishes with aluminum foil and keep in a growth chamber at 28 °C for 36 h. (Figure 2H).
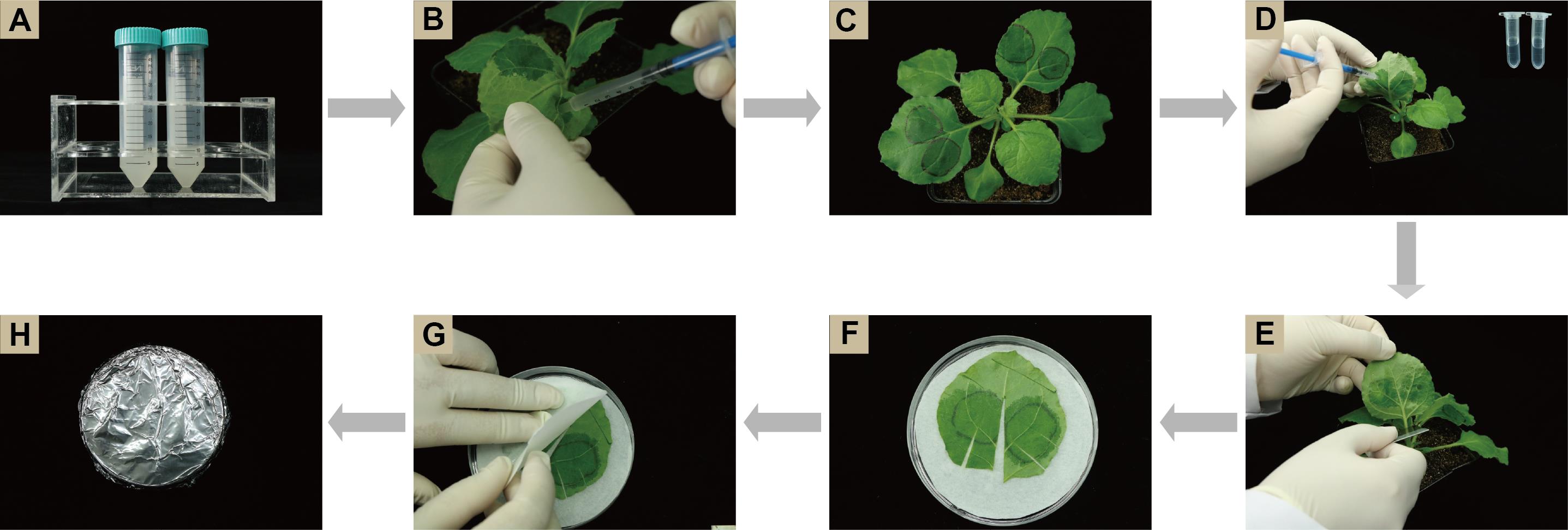
Figure 2. Illustration of the experimental procedure. (A) Resuspend bacterial solution; (B) Infiltrate bacterial mixture; (C) Mark infiltrated leaf areas; (D) Infiltrate ConA solution; (E) Detach infiltrated leaves; (F) Place detached leaf on pre-wetted filter paper; (G) Cover leaf with one more layer of pre-wetted filter paper; (H) Wrap Petri dish with aluminum foil.
Fluorescence Confocal Microscopy Imaging
Cut four leaf squares (0.5 × 0.5 cm) surrounding the infiltrated sites and mount in ddH2O water.
Examine the adaxial epidermal cells to visualize the subcellular localization of GFP-ATG8a and POI-mCherry with a Zeiss LSM 800 laser scanning confocal microscope (Carl Zeiss, https://www.zeiss.com).
Setup of confocal microscope: for GFP, excitation at 488 nm with an Argon laser and emission detection at 505-550 nm; For mCherry, excitation at 543 nm with an HeNe and emission detection at 585-615 nm. Use a laser strength of 4% and a pinhole of 1 airy unit for both fluorescence proteins.
Data analysis
As shown in Figure 3, numerous puncta decorated by autophagy marker GFP-ATG8a were readily detected in the vacuole when treated with ConA, a specific inhibitor of vacuolar-type ATPases (Dröse et al., 1993; Dettmer et al., 2006). Moreover, POI-mCherry labeled puncta accumulated in the vacuole, colocalizing with GFP-ATG8a upon ConA treatment, indicating the association of POI with autophagic vesicles. In contrast, no mCherry labeled puncta were detected in the vacuole regardless of ConA treatment. The colocalization assay with GFP-ATG8 based on the tobacco transient expression system shown here could be used for the identification of ATG components or autophagic cargoes, together with other approaches such as protein–protein interaction assays and ATG8 lipidation assay.

Figure 3. Representative confocal images showing the subcellular localization of POI-mCherry and GFP-ATG8a. N. benthamiana leaf epidermal cells were co-infiltrated with mCherry (A) or POI-mCherry (B) and GFP-ATG8a, and then analyzed with confocal microscopy 24 h after agroinfiltration followed by a 36-h incubation with 1 μM ConA or DMSO treatment (–ConA). AB: autophagic body; scale bar = 10 μm.
Recipes
Bacterial suspension buffer (must be freshly prepared before use)
10 mM MgCl2
100 µM Acetosyringone
10 mM MES (adjusted with 1 M KOH to pH = 5.8)
In ddH2O
ConA stock solution (0.5 mM)
Prepare stock solution by adding 230.9 μL DMSO to 100 µg ConA. Dispense the solution into 50 μL aliquots and store them at -20 °C or below.
MS-C liquid medium
4.3 g MS basal salt mixture
2 µM MES monohydrate
In 1 L ddH2O (adjusted with 1 M KOH to pH = 5.7)
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Guangdong Province (Grant 2022A1515011483) and South China Agricultural University Students’ Innovation and Entrepreneurship Training program (202110564052) to NL, and the National Natural Science Foundation of China (Grant 31970307) to FL and (Grant 31401906) to NL. The original paper in which this protocol was used is Luo et al. (2021; doi.org/10.1101/2021.06.11.448008).
Competing interests
The authors declare no conflicts of interest.
References
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18(3): 715-730.
- Dröse, S., Bindseil, K. U., Bowman, E. J., Siebers, A., Zeeck, A. and Altendorf, K. (1993). Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32(15): 3902-3906.
- Luo, N., Shang, D., Tang, Z., Huang, X., Tao, L.-Z., Liu, L., Gao, C., Qian, Y., Xie, Q. and Li, F. (2021). Engineered Aim-based selective autophagy to degrade proteins and organelles. 2021.2006.2011.448008.
- Marshall, R. S. and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol 29(69): 173-208.
- Suttangkakul, A., Li, F., Chung, T. and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761-3779.
- Tanz, S. K., Castleden, I., Small, I. D. and Millar, A. H. (2013). Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front Plant Sci 4: 214.
- Zhuang, X., Wang, H., Lam, S. K., Gao, C., Wang, X., Cai, Y. and Jiang, L. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25(11): 4596-4615.
Article Information
Publication history
Accepted: Jul 4, 2022
Published: Aug 20, 2022
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mai, J., Shang, D., Li, F. and Luo, N. (2022). Colocalization Assay with Fluorescent-tagged ATG8 Using a Nicotiana benthamiana-based Transient System. Bio-protocol 12(16): e4486. DOI: 10.21769/BioProtoc.4486.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Fluorescence
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







