- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
THRIFTY—A High-throughput Single Muscle Fiber Typing Method Based on Immunofluorescence Detection
(*contributed equally to this work) Published: Vol 13, Iss 10, May 20, 2023 DOI: 10.21769/BioProtoc.4678 Views: 397
Reviewed by: Alessandro DidonnaMarco Pagliusi Jr.Xin Xu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
In situ Microinflammation Detection Using Gold Nanoclusters and a Tissue-clearing Method
Fayrouz Naim [...] Masaaki Murakami
Apr 5, 2023 702 Views
E15.5 Mouse Embryo Micro-CT Using a Bruker Skyscan 1172 Micro-CT
Elena Astanina [...] Federico Bussolino
May 5, 2023 310 Views
Princeton RAtlas: A Common Coordinate Framework for Fully cleared, Whole Rattus norvegicus Brains
Emily Jane Dennis [...] Carlos D. Brody
Oct 20, 2023 567 Views
Abstract
Skeletal muscle consists of a mixture of fiber types with different functional and metabolic characteristics. The relative composition of these muscle fiber types has implications for muscle performance, whole-body metabolism, and health. However, analyses of muscle samples in a fiber type–dependent manner are very time consuming. Therefore, these are often neglected in favor of more time-efficient analyses on mixed muscle samples. Methods such as western blot and myosin heavy chain separation by SDS-PAGE have previously been utilized to fiber type–isolated muscle fibers. More recently, the introduction of the dot blot method significantly increased the speed of fiber typing. However, despite recent advancements, none of the current methodologies are feasible for large-scale investigations because of their time requirements. Here, we present the protocol for a new method, which we have named THRIFTY (high-THRoughput Immunofluorescence Fiber TYping), that enables rapid fiber type identification using antibodies towards the different myosin heavy chain (MyHC) isoforms of fast and slow twitch muscle fibers. First, a short segment (<1 mm) is cut off from isolated muscle fibers and mounted on a customized gridded microscope slide holding up to 200 fiber segments. Second, the fiber segments attached to the microscope slide are stained with MyHC-specific antibodies and then visualized using a fluorescence microscope. Lastly, the remaining pieces of the fibers can either be collected individually or pooled together with fibers of the same type for subsequent analyses. The THRIFTY protocol is approximately three times as fast as the dot blot method, which enables not only time-sensitive assays to be performed but also increases the feasibility to conduct large-scale investigations into fiber type specific physiology.
Graphical Overview

Graphical overview of the THRIFTY workflow. Cut off a small segment (0.5 mm) of an individually dissected muscle fiber and mount it onto the customized microscope slide containing a printed grid system. Using a Hamilton syringe, fixate the fiber segment by applying a small droplet of distilled water on the segment and let it fully dry (1A). The remaining large segment of the fiber should be placed in the corresponding square on a black A4 paper (1B). Once the microscope slide has been fully mounted with fiber segments, submerge the slide in a polypropylene slide mailer (illustrated as a Coplin jar in the figure) containing acetone to permeabilize the fiber segments. Thereafter, incubate the slide with primary antibodies targeting MyHC-I and MyHC-II. Following washes in PBS solution, incubate the slides with fluorescently labeled secondary antibodies, wash again, and mount with a cover glass and antifade reagent (2). Identification of fiber type can be performed using a digital fluorescence microscope (3), whereafter the remaining pieces of the fiber segments (large) are pooled together according to their fiber type or individually collected for experiments on single fibers (4). Image modified from Horwath et al. (2022).
Background
Skeletal muscle consists of a mixture of muscle fiber types, unique in their contractile and metabolic properties, as well as in their proteomic profile (Murgia et al., 2021). At a whole-muscle level, the relative abundance of the fiber types determines whether the muscle is primarily adapted to low-intensity repetitive activities or to short bursts of high-intensity contractions. The relative abundance of fiber types has also been linked to a number of physiological outcomes at a whole-body level, such as obesity and weight loss (Tanner et al., 2002), glucose-stimulated insulin secretion (Blackwood et al., 2022), and risk of cardiovascular disease (Karjalainen et al., 2006). In addition, many reports have demonstrated that skeletal muscle responds to acute exercise stimuli in a fiber type–dependent manner (Koopman et al., 2006; Tannerstedt et al., 2009; Kristensen et al., 2015). Similar observations have been reported for muscle adaptations in response to long-term training regimens, such as muscle fiber hypertrophy, improved glucose transport, and increased mitochondrial content (Daugaard et al., 2000; Verdijk et al., 2009; Skelly et al., 2021). Understanding the molecular intricacies of the different fiber types of skeletal muscle is therefore important for both disease prevention and physical performance.
Fiber type–specific analyses of isolated muscle fibers are typically associated with a tedious and time-consuming workflow, including isolating and fiber typing of hundreds to thousands of individual fibers prior to the experimental analysis. Even though the process of fiber type identification was recently improved by the introduction of the dot blot protocol (Christiansen et al., 2019), this method is still limited by the time required to fiber type large quantities of muscle fibers. The dot blot protocol also requires the entire muscle fiber to be denatured, thus limiting the number of analytical applications that can be performed on the same sample lysate. Therefore, we set out to develop the THRIFTY (high-THRoughput Immunofluorescence Fiber TYping) method to further facilitate the workflow associated with fiber type identification of single muscle fibers. The THRIFTY protocol presented herein will drastically reduce the time consumption of fiber type identification, thus allowing researchers to more easily capture a representative portion of the total muscle fiber pool. Our method also allows for experimental techniques in which a large sample mass is required to obtain proper measurement resolution, e.g., analyses of muscle protein synthesis using stable isotope tracers.
Materials and Reagents
Microscope slides (VWR, catalog number: 631-1554)
Cover slips 24 × 50 mm (VWR, catalog number: 631-1574)
Test tubes, soda glass 40 × 8 mm (VWR, catalog number: 212-0011)
Versilic peroxide-cured silicone stoppers (Lab Pure, catalog number: 263006-50)
CryoPure tubes 1.2 mL (Sarstedt, catalog number: 72.377)
Silica gel, SiO2 (VWR, catalog number: 83001.260)
Milli-Q ultrapure water
Acetone (C3H6O) (Fisher Scientific, catalog number: 1000220500)
Phosphate buffered saline (PBS) tablets (Fisher Scientific, catalog number: 10388739)
TritonTM X-100 (Sigma-Aldrich, catalog number: T8787)
Normal goat serum (NGS) 10% (Thermo Fisher Scientific, catalog number: 50062Z), store at 4 °C
Mouse anti-myosin heavy chain isoform I (DSHB, catalog number: BA-F8-c), store at 4 °C
Mouse anti-myosin heavy chain isoform II (DSHB, catalog number: SC-71-c), store at 4 °C
Alexa Fluor goat anti-mouse IgG2b 488 (Thermo Fisher Scientific, catalog number: A-21141), store at 4 °C in the dark
Alexa Fluor goat anti-mouse IgG1 647 (Thermo Fisher Scientific, catalog number: A-21240), store at 4 °C and in the dark
ProlongTM Gold antifade mountant (Thermo Scientific, catalog number: P36934)
Triton 10% stock solution (see Recipes)
Phosphate buffered saline (PBS) solution (see Recipes)
Primary antibody solution (see Recipes)
Secondary antibody solution (see Recipes)
Equipment
Freeze-dryer system (Heto FD 1.0 cooling unit, Edwards VacuumTM nXDS6i Vacuum Pump and Vacuubrand VAP5 Vacuum Gauge)
Climate controlled room (<40% humidity)
VisiScope® Stereo microscope (VWR, catalog number: 630-3073)
Dumont forceps (FST, catalog number: 11252-00)
Needles (sharpened sewing needles glued to a wooden handle)
Scalpel (Swamm-Morton, carbon steel scalpel, catalog number: 11)
HamiltonTM syringe 10 μL (Fisher Scientific, catalog number: 203560)
Black A4 paper (Common office supply)
Set of micropipettes (Tacta®, mechanical pipettes 0.5–1,000 μL, Sartorius)
Pipette tips (Safetyspace®, filtered pipette tips 0.5–1,000 μL, Sartorius)
Vortex-Genie® (VWR, catalog number: 444-5900)
Magnetic stirrer (Heidolph Instruments, catalog number: 503-02000-00)
Polypropylene slide mailer (Histolab, catalog number: 05309)
Celena S fluorescence microscope (Logos Biosystems)
Celena S LED filter cube (EYFP Ex500/20, Em535/30; Logos Biosystems, catalog number: I10106)
Celena S LED filter cube (Cy5 Long Pass Ex620/60, Em665 lp; Logos Biosystems, catalog number: I10112)
Pen and paper
Software
Celena S Digital Imaging Software (Logos Biosystem)
Procedure
Prepare microscope slides and grid system on black and white A4 paper
Print grid on a microscope slide with solvent-resistant ink using any commercially available printing service with glassware-printing capabilities (Figure 1A).
Using a regular pencil and ruler, draw a grid system corresponding to the printed microscope slide on black A4 paper (gridded paper reusable; Figure 1B).
Using a word processing software (e.g., Microsoft Word), create a similar grid system as in the previous step and print on white A4 office paper for later use during step F6 (Figure 1C).
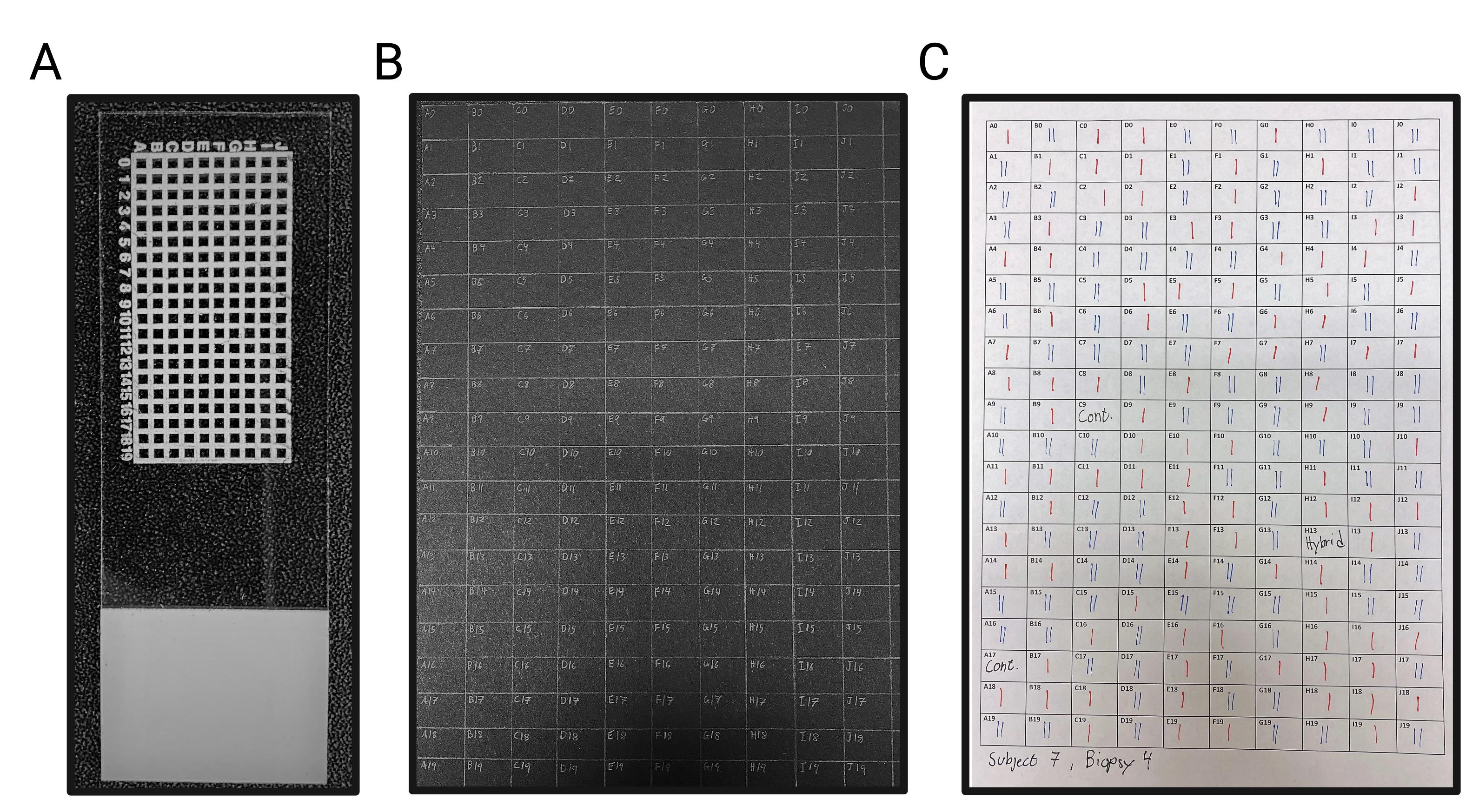
Figure 1. The coordinate grid system used throughout the THRIFTY protocol.A. Microscope slide with a custom coordinate grid system printed with solvent-resistant ink. Each square measures 1.05 × 1.05 mm, with a line thickness of 0.5 mm. The entire microscope slide measures 76 × 26 mm. B. A regular A4-sized black paper with a corresponding grid system for keeping track of the remainder of each muscle fiber. C. A matching grid system created in Microsoft Word and printed on regular A4 office paper. This paper is used to note the fiber type identity of each fiber segment when the microscope slide is visualized in the microscope. Type I and type II fiber segments are marked in red (I) and blue (II), respectively.
Isolate individual muscle fibers
Freeze-dry a muscle biopsy sample overnight and store at -80 °C. Before freeze-drying, start the cooling system connected to the vacuum pump and place the muscle sample in a cryotube with small holes in the lid to allow for evaporation. Place the sample in a plastic container (specific to the freeze-dryer) and put it in the -80 °C freezer. Once the plastic container has reached -80 °C, quickly remove it from the freezer and attach it to the freeze-dryer. The sample is then left to dry at a pressure below 0.03 mbar overnight.
Prior to dissection, thaw the biopsy sample to room temperature on silica gel for 30 min.
In a climate-controlled room (<40% humidity), manually isolate individual muscle fibers using needles and forceps under a stereomicroscope (Figure 2). Do this by first locating a fiber bundle of appropriate length, and then splitting the fiber bundle into smaller segments containing 2–10 fibers. Thereafter, fixate the bundle with the forceps and, using the needle, carefully remove individual fibers without breaking them.
Store dissected fibers in glass tubes sealed with silicone stoppers at -80 °C (it is important not to store fibers in plastic tubes, as it may be challenging to retrieve them from the test tubes prior to fiber type identification due to static electricity).

Figure 2. Manual isolation of individual freeze-dried muscle fibers. Isolation is performed under a stereomicroscope using Dumont forceps and custom needles created from sharpened sewing needles attached to a handle.
Mount fiber segment onto the microscope slide
Place the glass tubes with the fibers on silica gel for 30 min to allow thawing to room temperature.
Under a stereomicroscope, and using a needle and scalpel, cut a short segment (<1 mm) of an individual fiber (Figure 3A and 3B).
Place the fiber segment in the first square of the microscope slide (Figure 3C).
Add a droplet of distilled water (dH2O) onto the fiber segment using a Hamilton syringe (attachment occurs as the droplet slowly dries; Figure 3D and 3E).
Place the remaining (large) piece of the muscle fiber directly into the corresponding square of the gridded black A4 paper (Figure 3F).
Repeat steps C2–C5 until all squares of the microscope slide are filled with fiber segments.
Mount one to five microscope slides with fiber segments prior to staining.
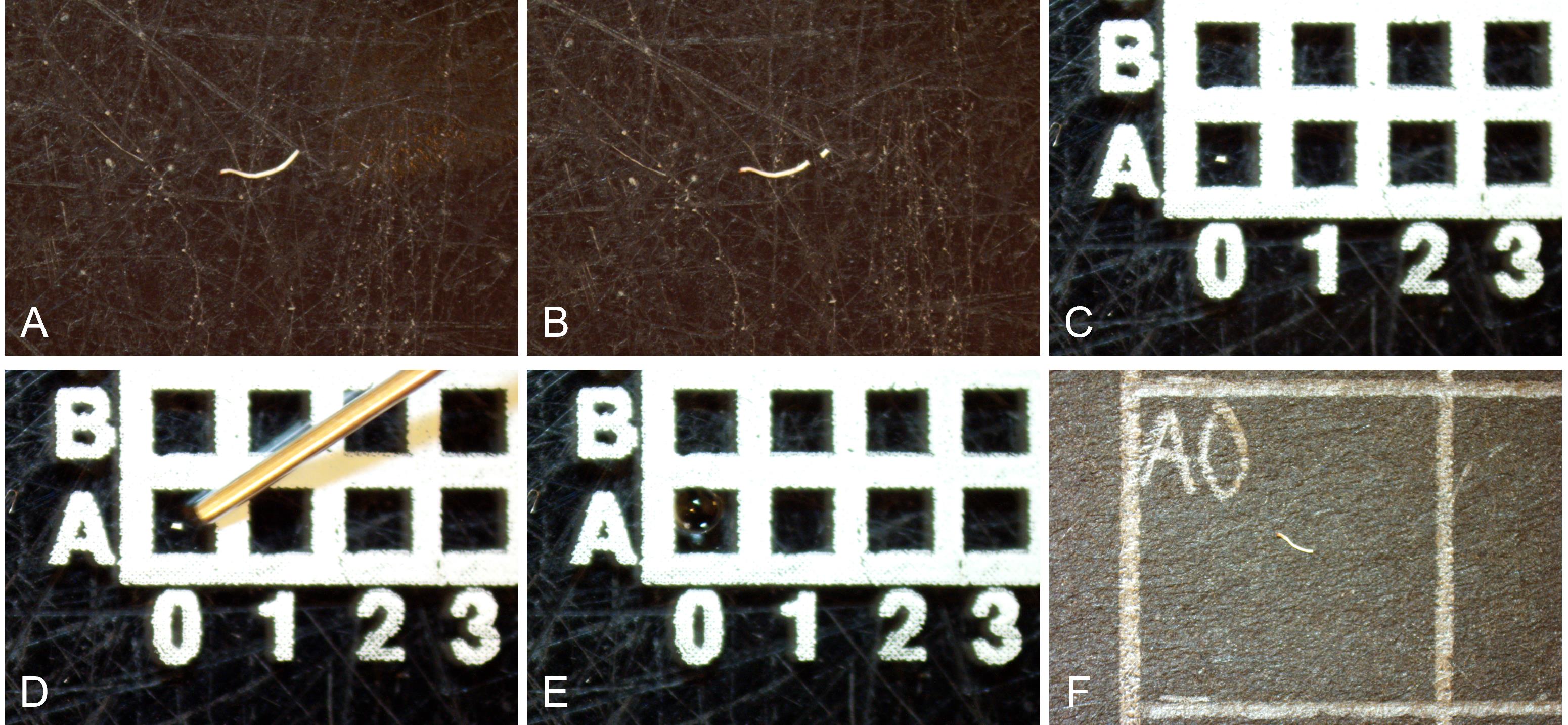
Figure 3. Mounting fiber segments and utilizing the coordinated grid system for fiber tracking. A. Isolate a fiber under the microscope. B. Cut the fiber into a large and a small fiber segment using a scalpel. C. Place the smaller fiber segment onto a square of the gridded microscope slide. D and E. Mount the small fiber end onto the microscope slide by adding a small droplet of distilled water onto it using a Hamilton syringe and letting it dry. F. Place the larger fiber end on the corresponding square of the gridded black A4 paper, to facilitate tracking which fiber end represents each of the remaining fiber pieces. All steps that require moving the fibers are done using needles, as muscle fibers generally stick to the needle due to static electricity.
Prepare stock solutions and the primary antibody solution
Prepare a Triton 10% stock solution (see Recipes).
Prepare PBS solution (see Recipes).
Prepare a primary antibody solution (see Recipes).
Staining protocol
Carefully submerge the mounted microscope slide into a polypropylene slide mailer filled with room temperature acetone and incubate for 3 min.
Let the microscope slides dry for approximately 3 min.
Carefully submerge the microscope slides in the primary antibody solution and incubate for 45 min at room temperature.
During incubation with primary antibodies, prepare secondary antibody solution (see Recipes; not recommended to reuse).
Wash the microscope slides three times for 5 min in cold PBS (4 °C) solution by carefully submerging them into mail sliders filled with PBS solution. The slides should be washed without any shaking movements to avoid possible risk of detachment.
Incubate the microscope slides in the secondary antibody solution at room temperature in the dark.
Wash the microscope slides again three times for 5 min in cold PBS (4 °C) solution in the dark.
Carefully dry excess PBS solution from the microscope slide and apply 2–3 droplets of the antifade mounting agent onto the fibers, followed by mounting onto a cover glass.
Let the antifade mounting agent dry for 15 min in the dark.
Fiber type identification in the Celena S microscope
Locate the first fiber segment using the light microscope setting (4× magnification).
Adjust the focus to clearly visualize the fiber segment.
Set the microscope to the following image settings: Light 100%, Gain (18 dB), and 40- and 10 ms, respectively, for the EYFP and Cy5 Long pass channels. Use the function pseudo-color to more easily detect the different fiber types. Here, we used green as a pseudo-color for the EYFP channel and red as a pseudo-color for the Cy5 Long pass channel. For simplicity, the channels will hereafter be referred to as green (EYFP) and red (Cy5 Long pass).
Control the fiber end in both the green and red channels.
Identify the fiber type using the following scheme: type I fibers are stained green, type II fibers are stained red, type I/II hybrid fibers are stained yellow, and more than one fiber segment of different colors in a square indicates a contamination, either because two adjacent fibers were not separated properly or because a residual fiber segment attached to the main fiber during the dissection process. See Figure 4 for an example.
Make a note of the identified fiber type for each fiber mounted on the microscope slide on the gridded white A4 paper.
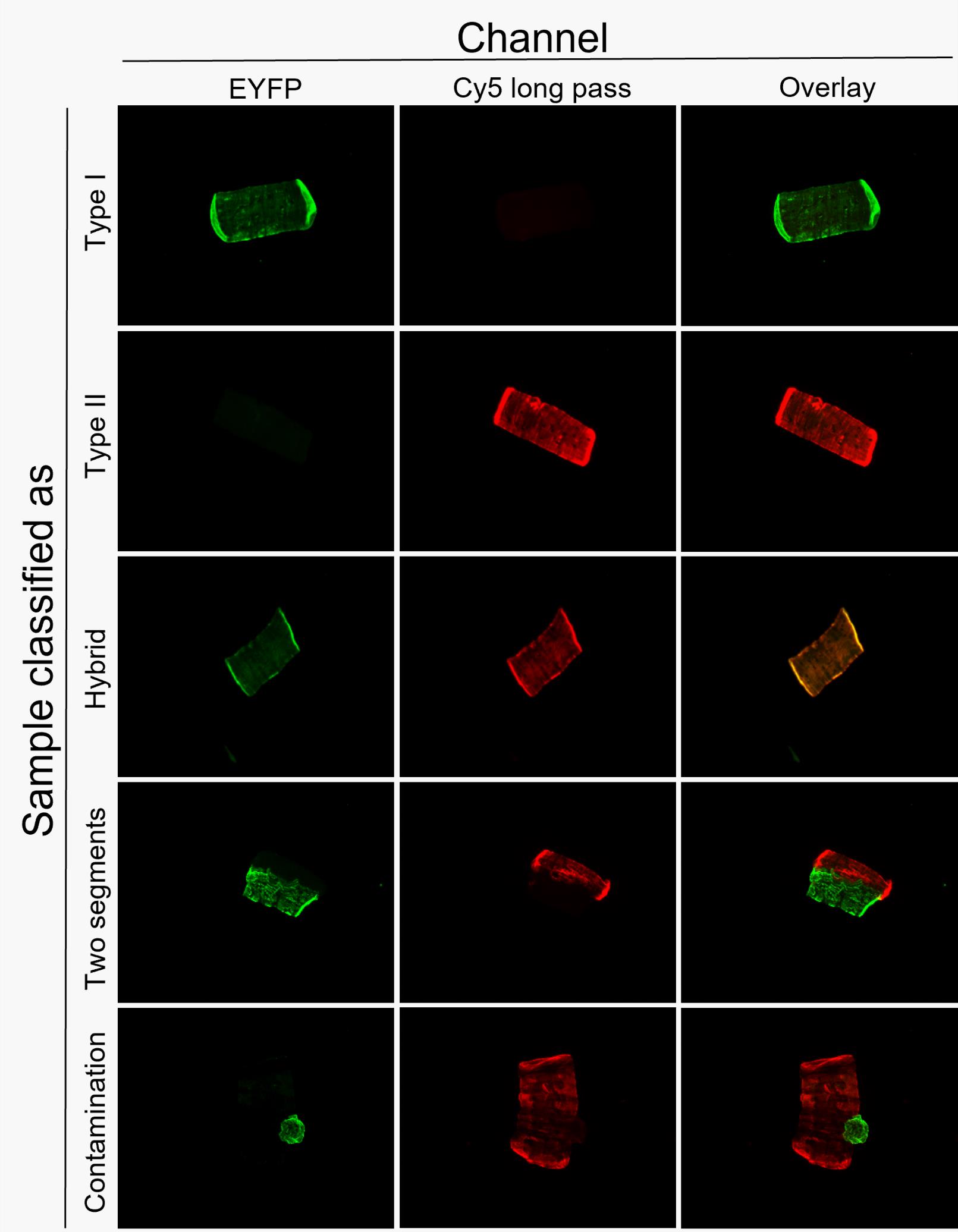
Figure 4. Examples of fiber type classification using the THRIFTY protocol. Fiber segments can either be classified as type I, type II, hybrid, two fiber segments, or a contamination. Image modified from Horwath et al. (2022).Locate the next fiber on the gridded microscope slide.
Repeat steps F4–F7 for the remainder of the fiber segments mounted onto the microscope slide.
Collect fibers individually or pool fibers according to their fiber type
The fiber type identity of each fiber placed on the black gridded A4 paper is now provided using the notes from the previous step.
This information can now be used to collect the fibers, either individually or pooled in line with their fiber type identify.
Proceed according to the requirement of the downstream analysis.
Notes
Printing service:
To create the customized glass slide containing a white grid system, we contacted a commercial printing company (Creative Reklam i Sverige AB, Ludvika, Sweden), which printed a 10 × 20 white grid system using solvent-resistant ink with a line thickness of 0.5 mm. Each square in the grid measured 1.05 × 1.05 mm. However, if microscope systems other than the Celena S are to be used, these dimensions can be adjusted accordingly to optimize usability.
To minimize the risk of fiber segments detaching from the microscope slide during the staining procedure, antibody solutions and PBS solution should not be poured over the microscope slide when placed in the slide mailer. Instead, the microscope slide should be carefully submerged into a pre-filled mailer. This includes all incubation and washing steps.
For the same reason as described above, plastic slide mailers are preferred over ordinary Coplin jars to minimize the potential for micro-vibrations, which could cause the fiber segments to detach during staining.
The authors strongly advise users to determine fiber type identity on the basis of the signal obtained in both the green and red channels (i.e., strong signal in the green channel and no signal in the red channel), rather than classifying a fiber segment on the basis of only a single channel (i.e., strong signal in the green channel). This is due to the large risk of misclassifying hybrid fibers, dual fiber segments, or contaminated fibers using this approach.
If speed of the staining procedure is crucial, primary and secondary antibody incubations can be reduced to 10 min each (incubation in 37 °C) and PBS solution washes can be performed once for 5 min instead of three times for 5 min. This allows for a staining protocol of approximately 30 min with a staining quality close to the original protocol.
The protocol described here is for freeze dried muscle; however, the THRIFTY method also works on fresh, non-freeze-dried muscle tissue.
Recipes
Triton 10% stock solution
Add 25 mL of Triton X-100 into 225 mL of dH2O.
Stir gently using a magnetic stirrer as Triton X-100 otherwise tends to create a layer of foam.
Store at room temperature or 4 °C.
Phosphate buffered saline (PBS) solution
Using a magnetic stirrer, dissolve two PBS tablets in 1 L of dH2O.
Store at 4 °C.
Primary antibody solution
Mix 6 mL of NGS with 4.8 mL of PBS solution.
Add 1.2 mL of Triton 10% stock into the solution; do not vortex yet.
As the antibody concentration varies between different batches, calculate the amount of mouse anti-myosin heavy chain isoform I and mouse anti-myosin heavy chain isoform II antibodies required for 30 and 50 μg of the solution, respectively.
Add antibodies to the solution based on the calculations.
Vortex.
Store at 4 °C.
Primary antibody solution can be reused multiple times if kept at 4 °C between uses (over 20 uses over a period of several months).
Secondary antibody solution
Prepare this solution fresh every use, preferably during primary antibody incubation.
Add 1 mL of Triton 10% stock solution and 1 mL of NGS into 8 mL of PBS solution.
Add 10 μL of Alexa Fluor goat anti-mouse IgG2b 488 and Alexa Fluor goat anti-mouse IgG1 647, respectively.
Vortex.
Keep the secondary antibody solution in the dark. The solution can be stored at room temperature if prepared during primary antibody incubation prior to use.
Acknowledgments
This protocol is based on a previous article published in The Journal of Physiology (Horwath et al., 2022). The project was funded by project grants (P2020-0058, P2021-0173) and an Early Research Fellowship (No. D2019-0050), both from the Swedish National Council for Sport Science awarded to W.A.
Competing interests
The authors have no competing interests.
Ethics
All experiments in the development of this method were conducted on human skeletal muscle samples. The muscle samples used to develop this protocol were obtained from three ongoing research projects approved by the Swedish Ethical Review Authority (DNR 2017/2107–31/2, DNR 2017/2034–31/2, and DNR 2019-0038 1). All projects were in agreement with the declaration of Helsinki, and all human volunteers gave both their oral and written consent prior to enrollment in experiments.
References
- Blackwood, S. J., Horwath, O., Moberg, M., Pontén, M., Apró, W., Ekblom, M. M., Larsen, F. J. and Katz, A. (2022). Extreme Variations in Muscle Fiber Composition Enable Detection of Insulin Resistance and Excessive Insulin Secretion. J Clin Endocrinol Metab 107(7): e2729-e2737.
- Christiansen, D., MacInnis, M. J., Zacharewicz, E., Xu, H., Frankish, B. P. and Murphy, R. M. (2019). A fast, reliable and sample-sparing method to identify fibre types of single muscle fibres. Sci Rep 9(1): 6473.
- Daugaard, J. R., Nielsen, J. N., Kristiansen, S., Andersen, J. L., Hargreaves, M. and Richter, E. A. (2000). Fiber type-specific expression of GLUT4 in human skeletal muscle: influence of exercise training. Diabetes 49(7): 1092-1095.
- Horwath, O., Edman, S., Andersson, A., Larsen, F. J. and Apró, W. (2022). THRIFTY: a novel high-throughput method for rapid fibre type identification of isolated skeletal muscle fibres. J Physiol 600(20): 4421-4438.
- Karjalainen, J., Tikkanen, H., Hernelahti, M. and Kujala, U. M. (2006). Muscle fiber-type distribution predicts weight gain and unfavorable left ventricular geometry: a 19 year follow-up study. BMC Cardiovasc Disord 6: 2.
- Koopman, R., Zorenc, A. H., Gransier, R. J., Cameron-Smith, D. and van Loon, L. J. (2006). Increase in S6K1 phosphorylation in human skeletal muscle following resistance exercise occurs mainly in type II muscle fibers. Am J Physiol Endocrinol Metab 290(6): E1245-1252.
- Kristensen, D. E., Albers, P. H., Prats, C., Baba, O., Birk, J. B. and Wojtaszewski, J. F. (2015). Human muscle fibre type-specific regulation of AMPK and downstream targets by exercise. J Physiol 593(8): 2053-2069.
- Murgia, M., Nogara, L., Baraldo, M., Reggiani, C., Mann, M. and Schiaffino, S. (2021). Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skelet Muscle 11(1): 24.
- Skelly, L. E., Gillen, J. B., Frankish, B. P., MacInnis, M. J., Godkin, F. E., Tarnopolsky, M. A., Murphy, R. M. and Gibala, M. J. (2021). Human skeletal muscle fiber type-specific responses to sprint interval and moderate-intensity continuous exercise: acute and training-induced changes. J Appl Physiol (1985) 130(4): 1001-1014.
- Tanner, C. J., Barakat, H. A., Dohm, G. L., Pories, W. J., MacDonald, K. G., Cunningham, P. R., Swanson, M. S. and Houmard, J. A. (2002). Muscle fiber type is associated with obesity and weight loss. Am J Physiol Endocrinol Metab 282(6): E1191-1196.
- Tannerstedt, J., Apro, W. and Blomstrand, E. (2009). Maximal lengthening contractions induce different signaling responses in the type I and type II fibers of human skeletal muscle. J Appl Physiol (1985) 106(4): 1412-1418.
- Verdijk, L. B., Gleeson, B. G., Jonkers, R. A., Meijer, K., Savelberg, H. H., Dendale, P. and van Loon, L. J. (2009). Skeletal muscle hypertrophy following resistance training is accompanied by a fiber type-specific increase in satellite cell content in elderly men. J Gerontol A Biol Sci Med Sci 64(3): 332-339.
Article Information
Publication history
Published: May 20, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Edman, S., Horwath, O. and Apró, W. (2023). THRIFTY—A High-throughput Single Muscle Fiber Typing Method Based on Immunofluorescence Detection. Bio-protocol 13(10): e4678. DOI: 10.21769/BioProtoc.4678.
Category
Molecular Biology > Protein > Expression
Cell Biology > Tissue analysis > Tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







