- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of scFv2H7-P18F3, a Bi-Modular Fusion Protein (BMFP) Targeting Human CD20
Published: Vol 13, Iss 10, May 20, 2023 DOI: 10.21769/BioProtoc.4682 Views: 470
Reviewed by: Victor TseRan ChenKuo-Ching MeiSuresh Kumar

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Heterologous Expression and High Degree Purification of the Restriction Endonuclease SauUSI
Vinayak Sadasivam Tumuluri and Kayarat Saikrishnan
Jan 5, 2022 2467 Views
Expression, Purification, and in vitro Enzyme Activity Assay of a Recombinant Aldehyde Dehydrogenase from Thermus thermophilus, using an Escherichia coli host
Kim Shortall [...] Tewfik Soulimane
May 5, 2022 1786 Views
Biosynthesis and Genetic Encoding of Non-hydrolyzable Phosphoserine into Recombinant Proteins in Escherichia coli
Philip Zhu [...] Richard B. Cooley
Nov 5, 2023 295 Views
Abstract
P18F3-based bi-modular fusion proteins (BMFPs), designed to re-direct pre-existing anti-Epstein-Barr virus (EBV) endogenous polyclonal antibodies towards defined target cells, demonstrated efficient biological activity in a mouse tumor model and could potentially represent a universal and versatile platform to develop novel therapeutics against a broad range of diseases. This protocol provides step-by-step instructions for expressing scFv2H7-P18F3, a BMFP targeting human CD20, in Escherichia coli (SHuffle®), and for purifying soluble proteins using a two-step process, namely immobilized metal affinity chromatography (IMAC) followed by size exclusion chromatography. This protocol can also be used for expression and purification of other BMFPs with alternative binding specificities.
Keywords: BMFPBackground
During the last decades, monoclonal antibody therapy has proven to be very effective for the treatment of many diseases such as cancers (Zahavi and Weiner, 2020), chronic inflammatory diseases (Voge and Alvarez, 2019), and certain diseases of infectious origin such as COVID-19 (Hwang et al., 2022). Nevertheless, the complex nature of immunoglobulins (Ig) (molecules of high molecular weight, with multiple chains and post-translational modifications impacting their effector functions) requires the use of demanding heterologous expression systems (mostly eukaryotic cell-based systems) for industrial production, which results in high manufacturing costs. To overcome this limitation, we recently conceptualized a novel immunotherapeutic approach and generated bi-modular fusion proteins (BMFPs), able to re-direct pre-existing anti-Epstein-Barr virus (EBV) endogenous polyclonal antibodies towards defined target cells, leading to their clearance. As a proof-of-concept, we have established that this strategy is efficient against cancer cells (Gamain et al., 2022). We have engineered and expressed, in Escherichia coli, a BMFP comprising a Fc-deficient binding moiety (scFv originating from the mouse IgG2b monoclonal antibody 2H7) targeting the human CD20 (huCD20) B-cell marker fused to a fragment of the EBV-P18 antigen (P18F3), which recruits circulating endogenous anti-P18F3 IgG in EBV+ individuals. We have shown that in vitro treatment of huCD20+ Burkitt’s lymphoma cells, in presence of plasma containing anti-P18F3 antibodies, elicits a significant activation of the antibody-dependent complement cascade and trigger FcγRIII-mediated activation of cellular pathways, leading to antibody-dependent cell-mediated cytotoxicity (Gamain et al., 2022). Furthermore, we have also demonstrated in a mouse tumor model that therapy performed with scFv2H7-P18F3 significantly leads to increased mice survival and full cancer remission in some animals (Gamain et al., 2022).
As P18F3-based BMFPs display efficient biological activity, they could potentially represent a universal and versatile platform to develop novel therapeutics against a broad range of diseases. This protocol provides step-by-step instructions for expressing and purifying scFv2H7-P18F3 in Escherichia coli (SHuffle®). It can also be used for expression and purification of other BMFPs with alternative binding specificities (Figure 1).
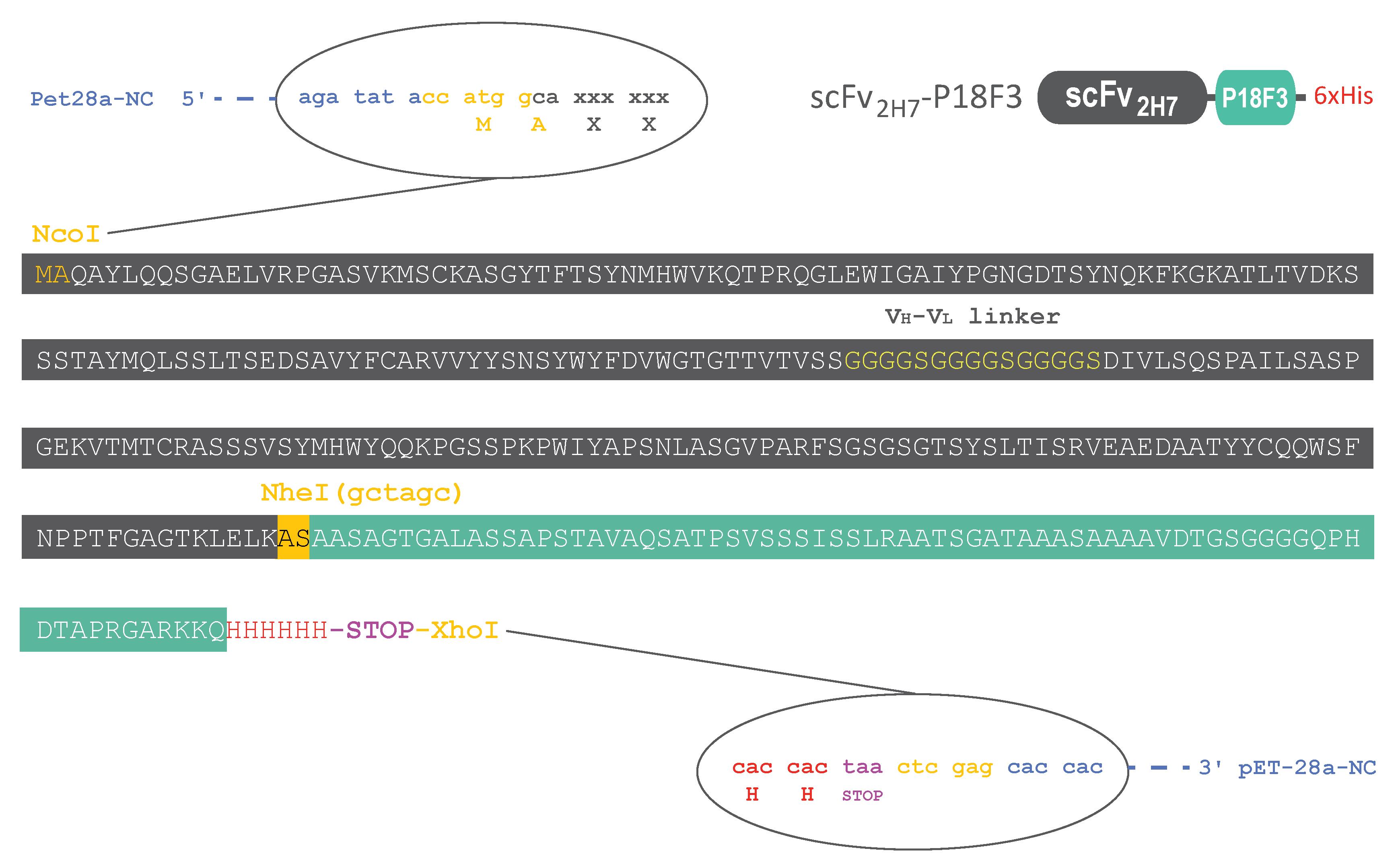
Figure 1. Standardized construct backbone for rapid and easy modulation of the binding moiety. The DNA sequence of scFv2H7-P18F3 was cloned into the pET-28a expression plasmid. The resulting amino acid sequence of scFv2H7 is displayed in dark gray, and the amino acid sequence of P18F3 is displayed in green. The recombinant protein possesses a C-terminal Histidine tag for easy purification and detection in functional assays. An alanine followed by a serine (AS) link scFv2H7 to the P18F3 polypeptide. When alternative binding specificity is desired, a new DNA sequence encoding for another binding moiety (devoid of Fc part) can be inserted between the NcoI and NheI restriction sites. We recommend using DNA sequences recoded for optimal E. coli codon usage, as it might ease the expression of the recombinant protein in SHuffle®.
Materials and Reagents
We describe here the exact materials and reagents that we use. Any equivalent materials and reagents may also be suitable for the procedure.
Materials
Filter tips PIPETMAN® DIAMOND 0.1–10 μL, 2–30 μL, 20–200 μL, 200–1,000 μL (Gilson, catalog numbers: F171203, F171303, F171503, F171703)
Sterile serology pipette; 2, 5, 10, 25, and 50 mL (Greiner, catalog numbers: 710160, 606180, 607107, 760180, 768160)
Expanded polystyrene ice bucket
Laboratory bottles with screw cap; 100 and 500 mL and 1 and 2 L (VWR, catalog numbers: 215-1592, 215-1594, 215-1595, 215-1596)
Petri dishes, polystyrene, 100 × 15 mm (Corning-Falcon, catalog number: 351029)
1.5 mL safe-lock Eppendorf microtubes (Eppendorf, catalog number: 0030121872)
Greiner culture tubes, 14 mL round bottom with snap vent cap (Merck, catalog number: Z617954)
Tube racks that can accommodate 1.5 (ø 11 mm), 15 (ø 17 mm), and 50 mL tubes (ø 30 mm)
L-shaped cell spreaders (Thermo Fisher Scientific, catalog number: SPCS01)
Tube with conical bottom; 15 and 50 mL (Corning-Falcon, catalog numbers: 352096, 352070)
Minisart high-flow syringe filter 0.2 μm pore size (Sartorius, catalog number: 16532)
Sterile syringes for single use; 1, 5, 10, and 60 mL (FisherbrandTM, Thermo Fisher Scientific, catalog numbers: 17161936, 15809152, 15819152, 15839152)
Cryotube 1.8 mL round bottom (Nunc, catalog number: 368632)
Aluminum foil
PES 0.2 μm filtration unit (Nalgene, catalog number: 564-0020)
High-resolution gel filtration Superdex® 200 Increase 10/300 GL column (Cytiva, catalog number: 28990944)
Reagents
Double-distilled water (ddH2O)
Ice
Super optimal broth with catabolite repression (SOC) medium (Thermo Fisher Scientific, catalog number: 15544034). Alternatively, SOC medium could be prepared according to the recipe provided in the Recipe section
SHuffle® T7 express competent E. coli (New England BioLabs, catalog number: C3029J)
NucleoSpin Plasmid QuickPureTM Kit (Macherey-Nagel, catalog number: 740615)
YPD (YEPD) agar powder (MP Biomedicals, catalog number: 114001222)
Lysogeny broth (LB) powder (MP Biomedicals, catalog number: 113002022)
Kanamycin sulphate powder (Thermo Fisher Scientific, catalog number: 15160054)
Imidazole (Merck, catalog number: I2399)
Phosphate buffer saline (PBS) pH 7.2 (Merck, catalog number: D8537). Alternatively, PBS could be prepared according to the recipe provided in the Recipe section
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Merck, catalog number: I5502)
Sodium chloride (NaCl) (Merck, catalog number: S7653)
Trizma® hydrochloride (Tris-HCl) (Merck, catalog number: 93363)
Lysozyme from chicken egg white (Merck, catalog number: L6876)
Protease inhibitor cocktail tablets (cOmpleteTM) (Merck, catalog number: 11697498001)
HisTrap FF 1 mL (prepacked with pre-charged Ni Sepharose® 6 fast flow) (Cytiva, catalog number: 17531901)
Laemmli sample buffer 4× (Bio-Rad, catalog number: 161-0747)
XT reducing agent (Bio-Rad, catalog number: 161-0792)
4%–15% 10-well mini-protean TGX stain-free gel (Bio-Rad, catalog number: 4568084)
10× Tris/Glycine/SDS (TGS) buffer (Bio-Rad, catalog number: 161-0772)
Nitrocellulose membrane 0.2 μm (Trans-Blot Turbo Transfer Pack) (Bio-Rad, catalog number: 1704158)
Western blotting detection reagents (ECLTM Prime) (Cytiva, catalog number: RPN2232)
Tryptone (Merck, catalog number: T2559)
Yeast extract (Merck, catalog number: Y1625)
KCl (Merck, catalog number: P9333)
MgCl2 (Merck, catalog number: M2670)
MgSO4 (Merck, catalog number: 63138)
Glucose (Merck, catalog number: G7528)
Centrifugal concentrator Amicon Ultra-4 (10 kDa MWCO) (Merck, catalog number: UFC8010)
Glycerol ≥99% (Merck, catalog number: G6279)
LB medium (1×) (see Recipes)
SOC medium (1×) (see Recipes)
Kanamycin stock solution (50 mg/mL) (see Recipes)
50% glycerol solution (see Recipes)
Bacteria resuspension solution/IMAC running buffer (see Recipes)
IMAC elution buffer (see Recipes)
Tris buffer saline (TBS) (see Recipes)
Tris buffer saline tween (TBST) (see Recipes)
Western blot blocking solution (TBS-5% milk) (see Recipes)
Western blot antibody dilution solution (TBST-5% milk) (see Recipes)
Phosphate buffer saline (PBS) (see Recipes)
Equipment
Pipette PIPETMAN® 0.1–10 μL, 2–20 μL, 20–200 μL, 100–1,000 μL (Gilson, catalog numbers: FA10002M, FA10003M, FA10005M, FA10006M)
Macroman® pipettor (Gilson, catalog number: F110120)
Borosilicate glass narrow neck Erlenmeyer flasks, 50 and 500 mL and 3 L (FisherbrandTM, Thermo Fisher Scientific, catalog numbers: 15499093, 15439103, 15469103)
Measuring glass cylinders, 100 and 500 mL and 1 and 2 L (VWR, catalog numbers: 612-3836, 612-3838, 612-3839; 612-3840)
Vertical gel electrophoresis system (PROTEAN Tetra cell) (Bio-Rad, catalog number: 1658004)
Autoclave (to be run at 120 °C for 20 min, under saturated steam ≥ 15 psi)
Water bath allowing precise temperature adjustment (range: 37–42 °C)
Shaking incubator allowing precise temperature adjustment (range: 20–37 °C), such as Multitron Standard incubator (Infors HT)
Incubator chamber allowing precise temperature adjustment (range: 30–37 °C)
Spectrophotometer allowing OD monitoring at 600 nm, such as BioSpectrometer® Basic (Eppendorf)
Standard fridge and freezer (-20 °C)
Ultra-low temperature freezer (-80 °C)
Refrigerated centrifuge, such as versatile 5810 series centrifuge (Eppendorf)
pH meter
Magnetic stirrer with magnetic stir bars (VWR, catalog number: 442-1271)
EmulsiFlex-C5 Homogenizer (Avestin) with air supply (recommended incoming pressure 120 psi/8 bar)
Vacuum pump
Power supply (Power PAC 300) (Bio-Rad, catalog number: 164-5050)
ChemiDocTM MP imaging system (Bio-Rad)
Rocking platform
Trans-Blot Turbo transfer system (Bio-Rad)
Automated liquid chromatography system, such as ÄKTA purifier 10 (Cytiva)
Chromatography refrigerator, such as Mediline LKPv 6522 (Liebherr)
Procedure
Expression of scFv2H7-P18F3
Antibody fragments such as scFv or VHHs (also known as nanobodies) possess structuring disulfide bonds mandatory for their functionality. In bacteria, disulfide bound formation requires translocation of proteins to the periplasmic compartment of the cell via an N-terminal signal peptide (Denoncin and Collet, 2013). Therefore, expression of disulfide-bonded recombinant proteins in standard E. coli–based systems usually leads to low production yields. Technological advances now enable overcoming this constraint by using mutant E. coli strains (such as SHuffle®), capable of promoting disulfide bound formation in the cytoplasm, leading to more efficient folding of recombinant proteins, improved activities, and increased production yields (Lobstein et al. 2012).
Day 1. Preparation of selection plates containing LB and agar supplemented with kanamycin (LB–agar K+) and of LB K+ expression medium
For 10 plates, weigh 2.25 g of agar powder (15 g/L) and 3.75 g of LB powder (25 g/L). Dissolve in 150 mL of ddH2O in a 250 mL autoclavable glass bottle. Put the lid back on the bottle without fully screwing it to allow pressure released during autoclaving.
Autoclave at 120 °C for 20 min, under saturated steam ≥ 15 psi.
Thaw a tube of kanamycin (stock solution at 50 mg/mL) on ice.
Once the autoclave cycle is done, let the bottle cool down to approximately 50 °C (at this temperature, you should be able to maintain your hand on the glass bottle for a few seconds).
Add 150 μL of kanamycin into the bottle and mix well to obtain LB–agar K+ (final concentration: 50 μg/mL).
Distribute 15 mL of LB–agar K+ into 10 Petri dishes (100 × 15 mm) (Figure 2A).
Let the plates cool down with the lid slightly open, until the LB–agar K+ is totally solidified and dried (Figure 2B).
Once the plates are dried, they can be stored at 4 °C (with the LB–agar K+ layer facing the top) for up to two weeks (Figure 2C).
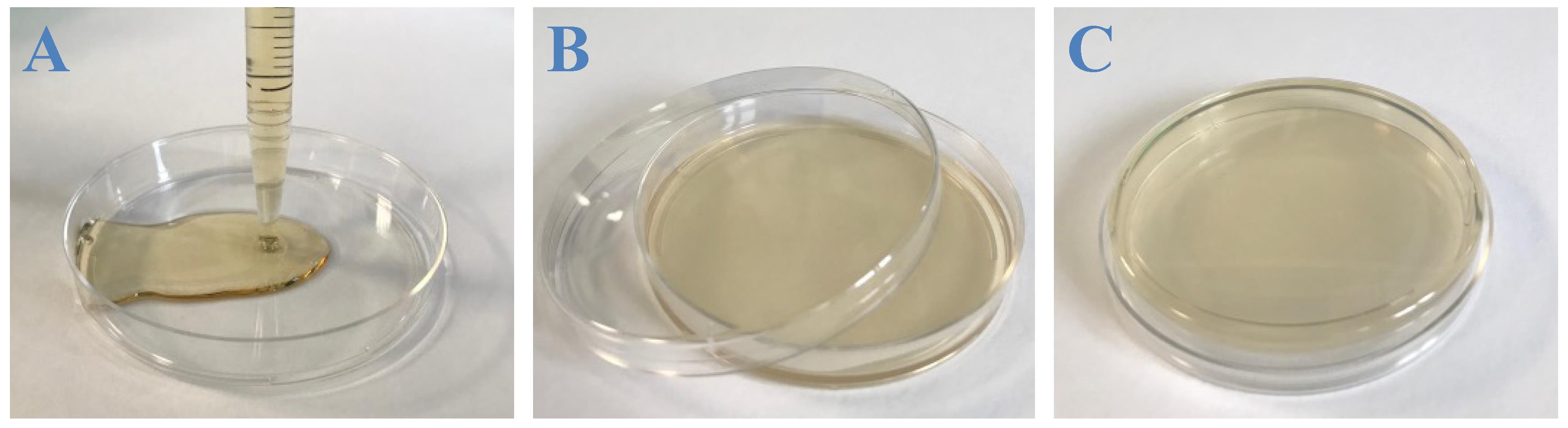
Figure 2. Casting LB–agar K+. A. 15 mL of warm LB–agar K+ are poured into a Petri dish using a serological pipette. B. The plate is then let to dry. C. Once fully dried, the plate is turned upside down and can be stored at 4 °C for up to two weeks.
Day 2. Transformation of SHuffle® T7 express competent E. coli with pET-28a-NC-(scFv2H7-P18F3)
Thaw pET-28a-NC-(scFv2H7-P18F3) DNA on ice.
Thaw a tube of SHuffle® T7 express competent E. coli (50 μL) on ice.
Add 50 ng of pET-28a-NC-(scFv2H7-P18F3) DNA (5 μL from the 10 ng/μL stock solution) to the bacteria suspension and gently mix (without vortexing).
Leave the tube on ice for 30 min. Meanwhile, equilibrate a water bath at 42 °C.
Remove the tube from the ice and dip it for 30 s in the water bath (42 °C heat shock).
Place the tube back on ice for 5 min.
Add 950 μL of room temperature SOC medium to the tube and transfer the whole volume (1 mL) to a new Greiner culture tube. Put the lid back on the tube without clipping it to allow air to go in.
Place the tube in a suitable rack and shake vigorously for 1 h at 30 °C. Here, we use a Multitron Standard incubator and set the tray revolution at 250 rpm. Meanwhile, equilibrate five LB–agar K+ selection plates at 30 °C in an incubator chamber.
Stop the incubation of the transformation mixture and perform five 10-fold serial dilutions in 1.5 mL safe-lock Eppendorf tubes using SOC medium (Figure 3).
Spread 100 μL of each dilution onto LB K+ selection plates using a L-shaped cell spreader.
Incubate the plates [plates should be inverted (Figure 2C)] at 30 °C in an incubator chamber overnight (18–20 h) for SHuffle® optimal growth.
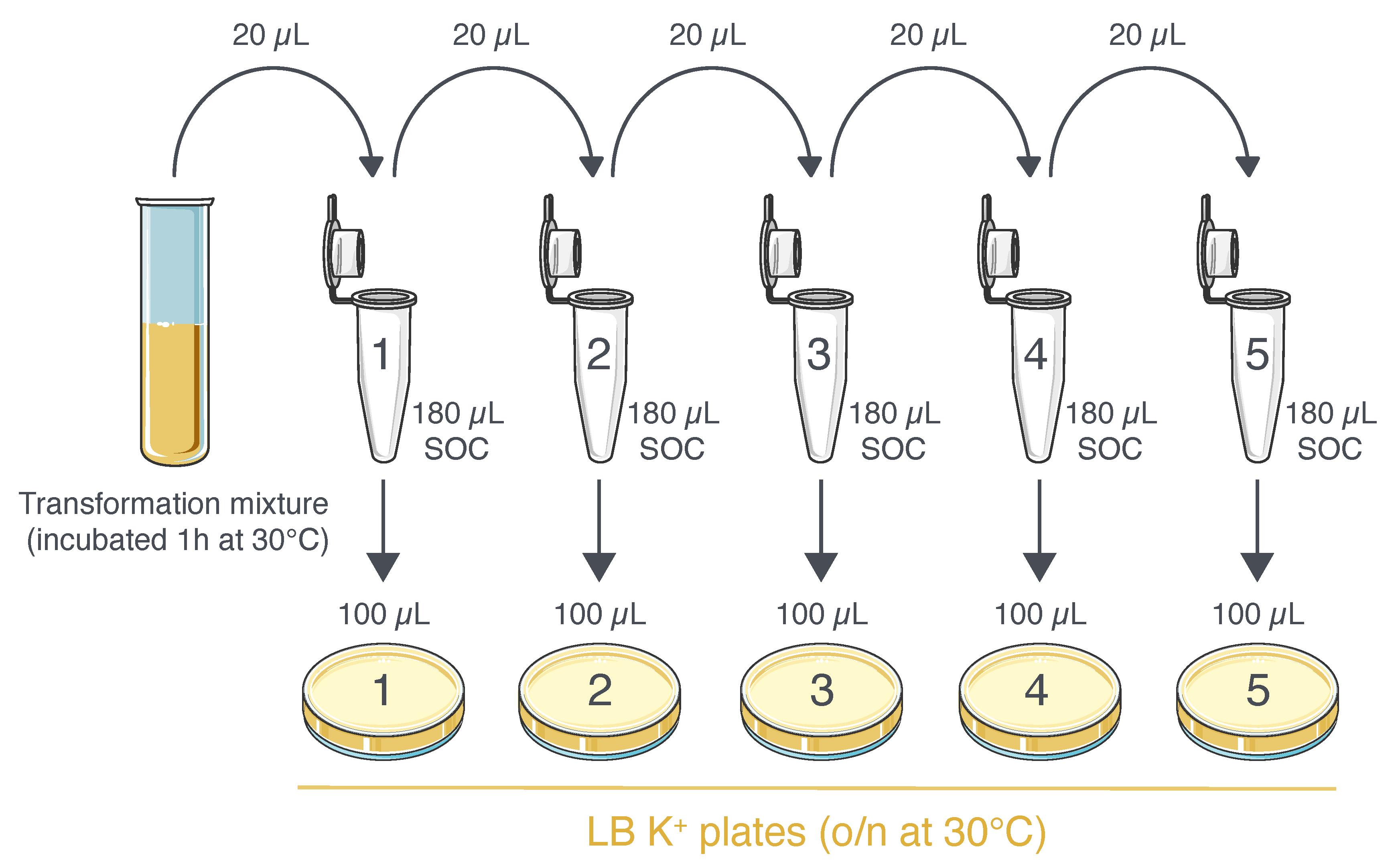
Figure 3. Plating procedure of competent E. coli transformed with pET-28a-NC-(scFv2H7-P18F3). The transformation mixture is subjected to serial dilutions in 1.5 mL tubes. 100 μL of each dilution is then used to seed five different LB K+ selection plates.
Day 3. Liquid bacteria culture from a single colony
Remove plates (plates 1–5) from the incubator and store them at 4 °C.
At the end of the day, thaw a tube of kanamycin (stock solution at 50 mg/mL) on ice.
Add 1 mL of the kanamycin stock solution to 1 L of LB medium.
Prepare a Greiner culture tube containing 1 mL of LB medium supplemented with kanamycin at 50 μg/mL (LB K+) and store the remaining LB K+ medium at 4 °C.
Pick up one single, isolated colony from one of the plates (plates 1–5) using a 200 μL clean tip (Figure 4A).
Place the tip inside the tube containing 1 mL of LB K+, put the lid back on the tube without clipping it, and incubate overnight at 30 °C with vigorous shaking (Figure 4B).
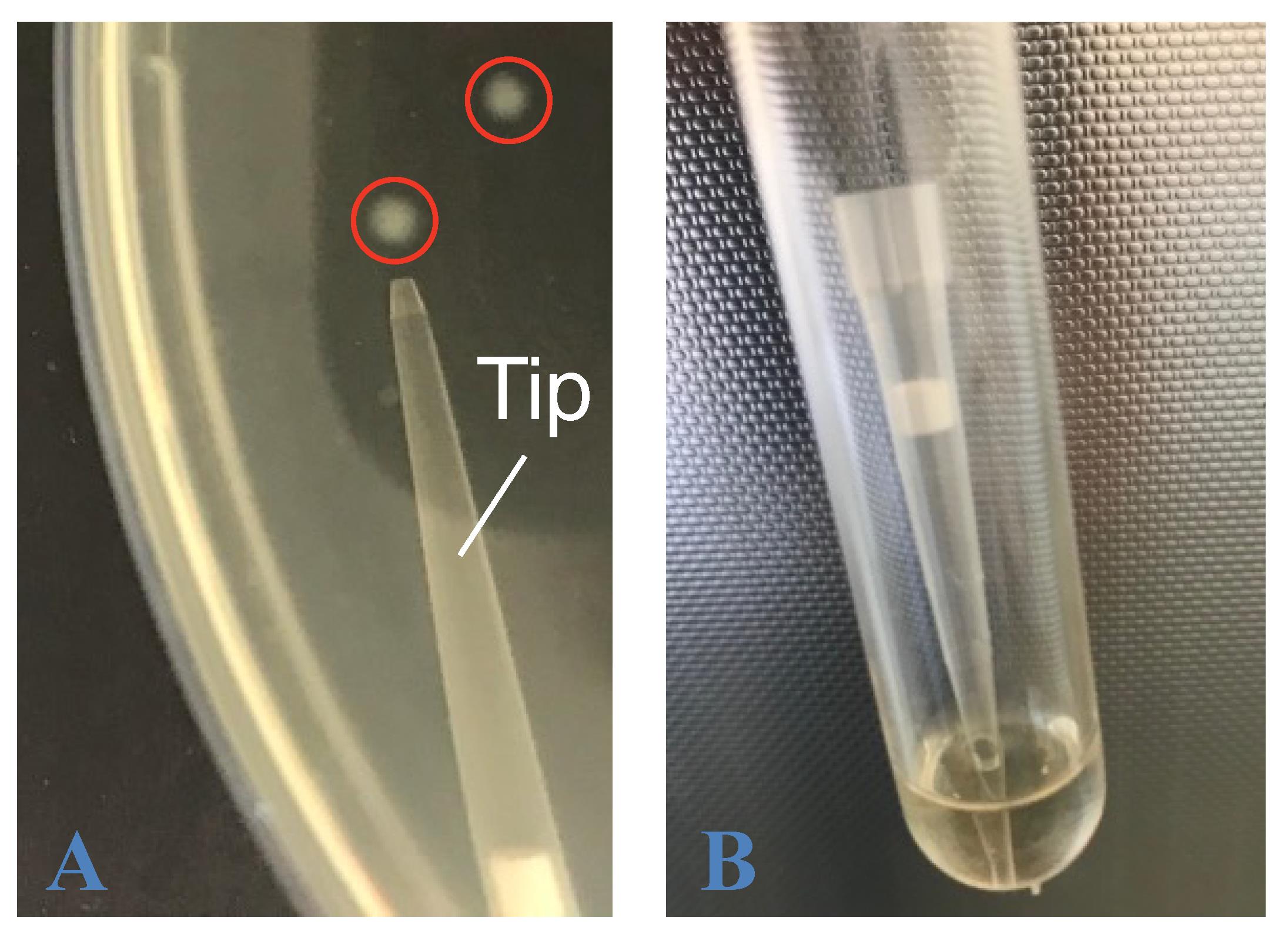
Figure 4. Picking up a single bacteria colony to inoculate an overnight liquid culture. A. A single bacteria colony is touched with a 200 μL pipette tip. B. The tip is then introduced in a tube containing 1 mL of LB K+. The tip can either be removed after a few seconds of swirling or left in the tube during overnight incubation at 30 °C.
Day 4. Bacteria culture scale up, induction of recombinant protein expression, glycerol stock preparation
Bacteria culture scaling up, induction of recombinant protein expression
Equilibrate 1 L of LB K+ medium at 30 °C in an incubator chamber.
Transfer 1 mL of the overnight culture into a 50 mL Erlenmeyer culture flask containing 9 mL of LB K+ medium. Cover the neck of the flask with aluminum foil (Figure 5A).
Incubate the flask with vigorous shaking at 30 °C for 3 h.
Transfer 10 mL of the culture into a 500 mL Erlenmeyer culture flask containing 90 mL of LB K+ medium. Cover the neck of the flask with aluminum foil (Figure 5B).
Incubate the flask with vigorous shaking at 30 °C for 3 h.
Transfer 100 mL of the culture into a 3 L Erlenmeyer culture flask containing 900 mL of LB K+ medium. Cover the neck of the flask with aluminum foil (Figure 5C).
Incubate the flask with vigorous shaking at 30 °C and check frequently (every 30 min) the optical density (OD600nm) of the culture using a spectrophotometer.
Thaw a tube of IPTG stock solution at 1 M on ice.
When OD reaches 0.8, collect 10 mL of the culture in a 15 mL Falcon tube (pre-induced sample) and keep it at room temperature until processing (see procedure in Figure 6).
Add 200 μL of IPTG from the stock solution into the flask (the final IPTG concentration is 0.2 mM) (Figure 5D).
Incubate the flask with vigorous shaking (250 rpm) at 20 °C overnight (14–16 h).
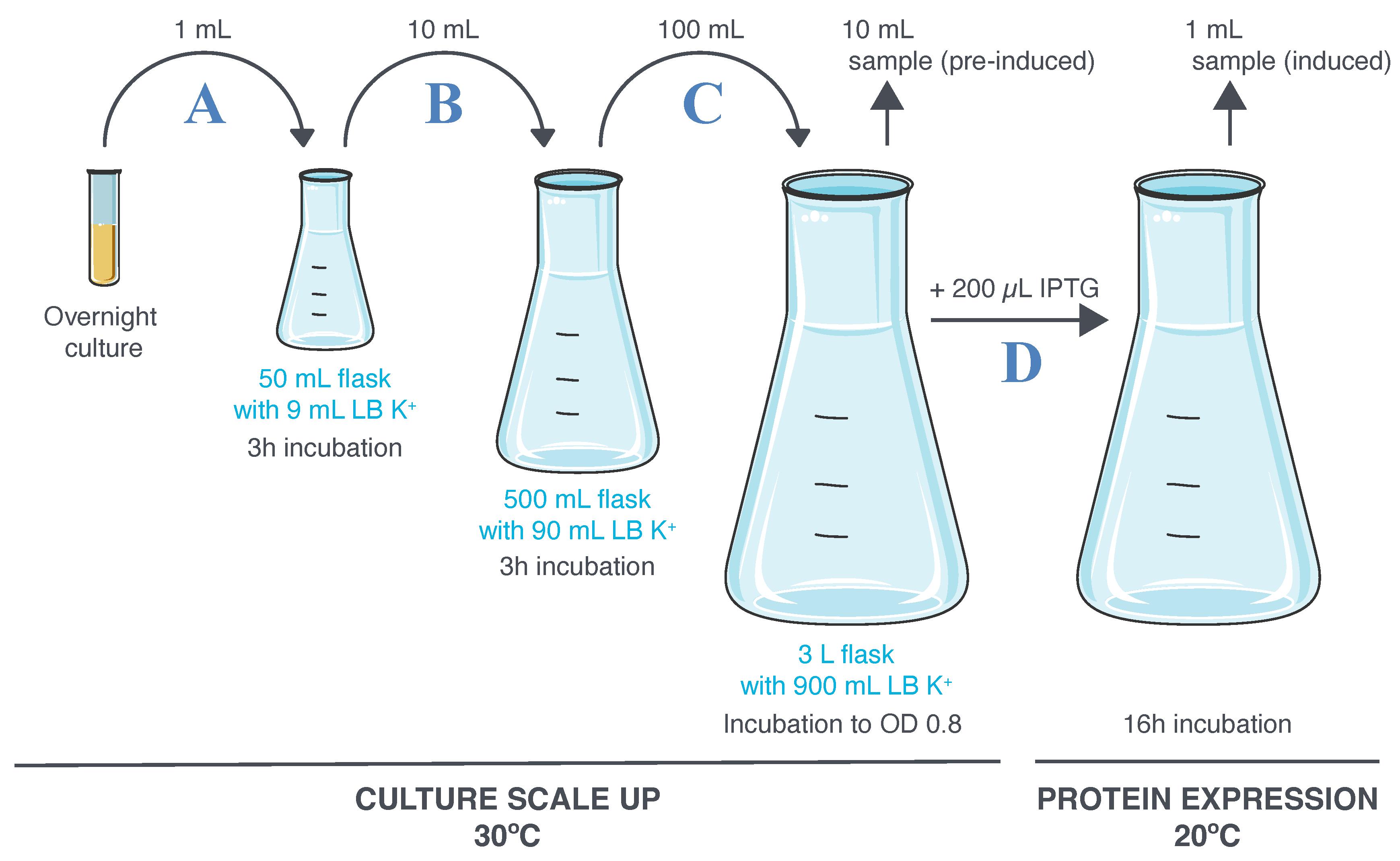
Figure 5. Culture scaling up and induction of scFv2H7-P18F3 expression. A–C. The overnight culture is expended following three consecutive 1:10 dilutions in LB K+ medium. D. A first aliquot of culture is sampled (pre-induced culture) before introduction of IPTG. A second aliquot of culture is sampled (IPTG-induced culture) after 16 h incubation at 20 °C.
Glycerol stock preparation
A glycerol stock of bacteria containing pET-28a-NC-(scFv2H7-P18F3) can be established to avoid cell transformation/plating steps each time a protein expression process is undertaken. This procedure should be performed right after sampling of the pre-induced culture aliquot.
Transfer 1 mL of the pre-induced culture sample (from Day 4, step 9) into a 1.5 mL safe-lock Eppendorf tube, centrifuge at 500× g for 20 min, remove the supernatant, and freeze the cell pellet at -20 °C until further analysis (Figure 6A).
Transfer 4 mL of culture from the pre-induced culture sample to a new 15 mL Falcon tube (Figure 6B).
Add 4 mL of 50% glycerol solution (see Recipes) into the tube and mix well by inverting up to 10 times.
Distribute the culture/glycerol mixture to eight cryotubes (1 mL per tube) and freeze them at -80 °C.
Sequencing of the plasmid at this stage is highly recommended.
Centrifuge the remaining 5 mL of culture sample (from Day 4, step 9) at 500× g for 10 min, discard the supernatant, and perform plasmid DNA isolation from the pelleted cells using a NucleoSpin Plasmid kit following the manufacturer’s instructions (Figure 6C).
The isolated plasmid DNA could then be sequenced to ensure suitability of the bacteria glycerol stock for new protein expression processes (glycerol stock validation).
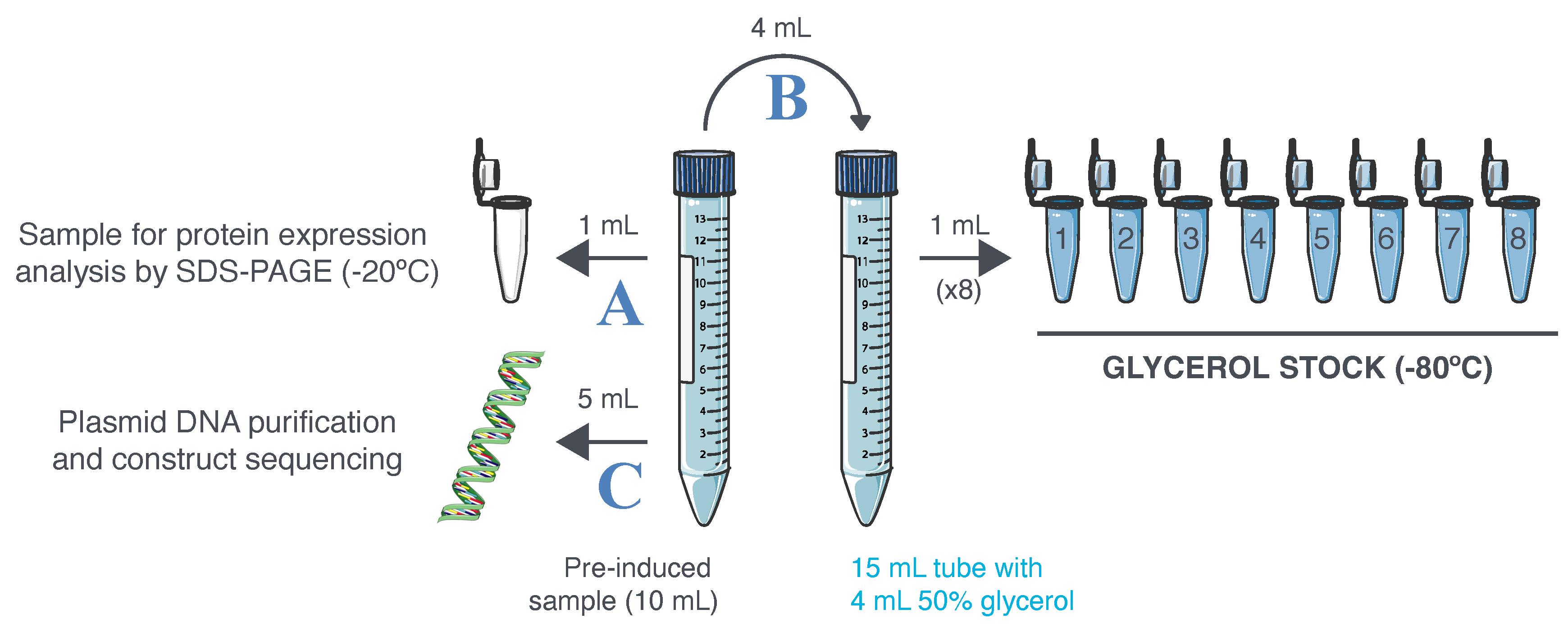
Figure 6. Glycerol stock preparation and validation. Ten milliliters of pre-induced sample are collected at Day 4, step 9. A. A first aliquot is frozen at -20 °C to serve as the non-induced culture sample during protein expression analysis by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). B. The pre-induced bacteria culture is mixed with 50% glycerol (v/v) and distributed into cryotubes. The cryotubes are then frozen at -80 °C to constitute a glycerol stock of bacteria. C. The leftover of the pre-induced sample is used to isolate plasmid DNA and ensure its correct sequence.
Addendum: Performing protein expression from a glycerol stock
A new protein expression process can be undertaken from a validated glycerol stock.
Prepare a Greiner culture tube containing 1 mL of LB medium supplemented with kanamycin (LB K+).
Remove a glycerol stock cryotube from -80 °C (do not let it thaw completely).
Immediately scrap the top of the frozen culture/glycerol mixture with a 200 μL clean tip.
Place the tip inside the tube containing 1 mL of LB K+, put the lid back on the tube without clipping it, and incubate with vigorous shaking overnight.
Put the glycerol stock cryotube back at -80 °C.
The following day, proceed with bacteria culture scale up and induction of recombinant protein expression as previously described (steps 1–11 of Day 4).
Day 5. Harvest of the IPTG-induced bacteria culture and assessment of protein expression
Culture harvest
Transfer 1 mL of the IPTG-induced culture sample into a 1.5 mL safe-lock Eppendorf tube, centrifuge at 500× g for 20 min, remove the supernatant, and freeze the cell pellet at -20 °C until further analysis by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Transfer the induced bacteria culture from the 3 L flask to four 500 mL centrifugation tubes.
Centrifuge the tubes at 500× g for 20 min at room temperature.
Discard the supernatant and resuspend each of the four bacteria pellets in 10 mL of bacteria resuspension solution (see Recipes).
Pool bacteria suspensions (4 × 10 mL) into a 50 mL Falcon tube and freeze at -80 °C until further use.
Assessment of protein expression
An assessment of protein expression is recommended before starting the purification processes. The following protocol describes how to analyze protein expression in whole culture samples by SDS-PAGE followed by western blotting. A finer analysis could also be undertaken on lysed bacteria by detecting the recombinant protein in the soluble as well as in the insoluble fractions*.
*Note: To prepare these fractions, the whole culture sample is centrifuged at 500× g for 20 min (4 °C) and the supernatant is discarded. The bacteria pellet is resuspended in PBS (same volume as discarded supernatant). Cells are lysed by repeated freeze/thaw cycles. The bacteria tube is put at -80 °C for 20 min, quickly thawed at 37 °C, and immediately put back at -80 °C for 20 min. Four to five freeze/thaw cycles might be needed for efficient cell lysis. The sample is then centrifuged at 15,000× g for 20 min (4 °C) and the supernatant, representing the soluble fraction is saved. PBS (same volume as the saved supernatant) is added to the insoluble material. Resuspension of the insoluble fraction can be achieved by dynamic pipetting followed by vigorous vortexing.
Thaw on ice the 1.5 mL safe-lock Eppendorf tubes containing the cell pellets of the pre-induced culture sample and of the IPTG-induced culture sample.
Resuspend cell pellets in 1 mL of bacteria resuspension solution.
Prepare two tubes, each containing 5 μL of 4× Laemmli sample buffer, 1 μL of XT reducing agent, and 14 μL of resuspended bacteria (non-induced and IPTG-induced).
Incubate tubes at 90 °C for 5 min.
Prepare a vertical electrophoresis system using a 4%–15% 10-well mini-protean TGX stain-free gel and TGS buffer as running buffer.
Load 20 μL of each sample into the wells and run the gel at 100 V until the blue migration front goes through the gel cast (approximately 2 h).
Uncast the gel and assess protein migration using the Stain Free technology on a ChemiDocTM MP imaging system (activation for 45 s) (Figure 7–left panel).
Transfer the gel onto a 0.2 μm nitrocellulose membrane using a Trans-Blot Turbo Transfer Pack and a Trans-Blot Turbo Transfer system (1.3 A, 25 V, 7 min).
Incubate the membrane in western blot blocking solution (TBS-5% milk; see Recipes) for 1 h at room temperature on a rocking platform.
Discard the blocking solution and incubate the membrane with an HRP-conjugated anti-His antibody (Penta His) diluted 1:10,000 in TBST-5% milk (see Recipes) for 1 h at room temperature on a rocking platform.
Discard the antibody solution and proceed with three consecutive membrane washes of 10 min each in TBST.
Perform a last membrane wash of 10 min in TBS.
Prepare 1 mL of western blotting detection solution (ECLTM Prime) by mixing 500 μL of Prime Luminol Enhancer Solution with 500 μL of Prime Peroxide Solution.
Drain excess of wash buffer from the membrane.
Add 1 mL of western blotting detection solution onto the membrane and incubate at room temperature for 5 min.
Drain excess of detection solution from the membrane.
Capture chemiluminescence signal using a ChemiDocTM MP imaging system (Figure 7–right panel).
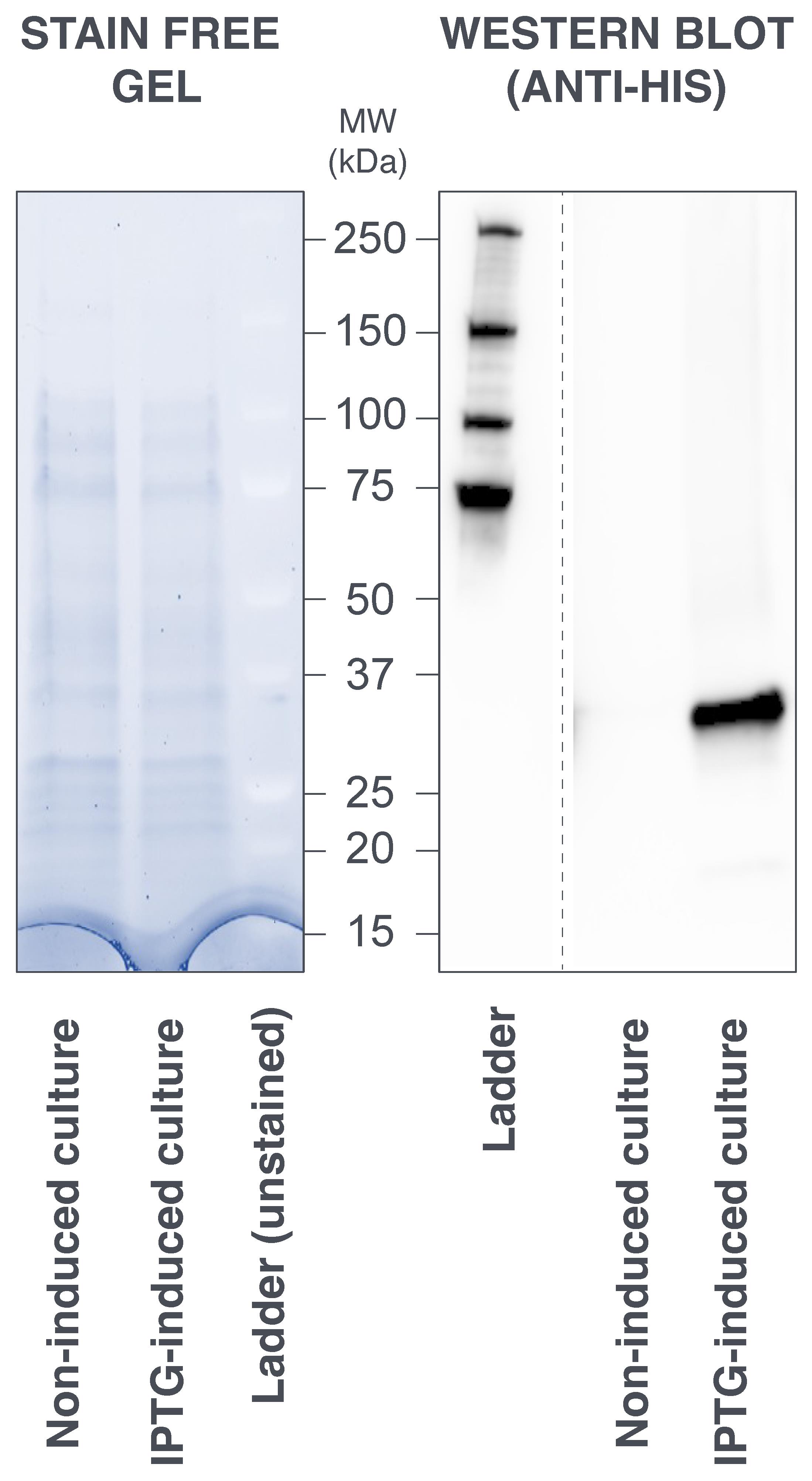
Figure 7. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of protein expression in whole bacteria culture. (Left panel) Migration profile of the non-induced and IPTG-induced culture samples. There is no apparent over-expression of scFv2H7-P18F3 (33.4 kDa) in the induced sample when the gel is subjected to Stain Free protein detection (artificial colors). (Right panel) Western blot analysis, performed using an anti-His antibody, revealed that scFv2H7-P18F3 is exclusively expressed in the IPTG-induced sample. MW: molecular weight. kDa: kilo Dalton.
Purification of scFv2H7-P18F3
Sample preparationThaw the bacteria suspension at room temperature.
Add 40 mg of lysozyme (1 mg/mL), mix well by vortexing, and keep the tube on ice.
Prime an EmulsiFlex-C5 homogenizer with bacteria resuspension solution at 4 °C.
Load 40 mL of bacteria suspension onto the homogenizer, perform bacteria lysis at a pressure of 14,500 psi, and collect the lysate. Repeat the lysis procedure two more times (total of three passages).
Dissolve one tablet of protease inhibitor in 1 mL of PBS.
Add 1 mL of protease inhibitor to the lysate.
Centrifuge the cell lysate at 20,000× g for 20 min at 4 °C in suitable centrifugation tubes.
Collect the supernatant in a 50 mL tube and filter it at 0.22 using a 50 mL filtering unit connected to a vacuum pump.
Let the vacuum pump on for a few minutes until the filtered solution is fully degassed (no more visible bubbles in the liquid). Indeed, lowering the pressure in the container will result in dissolving gas from the liquid. The sample may then be applied to a column without the risk of air bubbles forming in the column.
Purification by immobilized metal affinity chromatography (IMAC)
The following purification procedures (IMAC and size exclusion chromatography) are performed using a ÄKTA purifier 10 (Cytiva). The process can be easily adapted to other liquid chromatography systems. Working at temperatures ≤ 15 °C is highly recommended to minimize proteolysis.
Prime pump A (inlet A1) and pump B (inlet B1) with ddH2O.
Run the system without any column attached (flow: 1 mL/min) until pressure, conductivity, OD280nm, and OD215nm values are stable.
Pause the system and connect a HisTrap FF (1 mL) column to the system [set up the pressure alarm at total system pressure (~0.1 MPa) + column pressure limit (0.5 MPa)].
Wash the column with 5 mL of ddH2O (flow: 1 mL/min).
Pause the system and place the inlet A1 into a bottle containing 250 mL of IMAC running buffer (see Recipes) and the inlet B1 into a bottle containing 250 mL of IMAC elution buffer (see Recipes).
Equilibrate the column with 5 mL of IMAC running buffer (flow: 1 mL/min; gradient: 0% B).
Pause the system and place the inlet A1 into a 50 mL tube containing the sample to be processed.
Here, pump A is used as a sample pump. Using an independent, dedicated sample pump on newer systems is recommended.
Run the sample through the column in IMAC running buffer, 10 mM imidazole (flow: 1 mL/min; gradient: 2% B) (Figure 8).
Save the flowthrough using a fraction collector.
Pause the system and place the inlet A1 into the bottle containing IMAC running buffer (clean inlet with ddH2O beforehand).
Wash the column with 15 mL of IMAC running buffer, 40 mM imidazole (flow: 1 mL/min; gradient: 8% B) (Figure 8). When purifying a novel type of BMFP, we recommend performing a scouting run prior to large scale purification to establish the best percentage of B to be used during the washing step (the protein of interest should not elute from the column).
Save the wash fraction using the collector.
Elute the column-bound material with 5–7 mL of IMAC running buffer, 250 mM imidazole (flow: 1 mL/min; gradient: 50% B) (Figure 8).
Collect the eluted fraction.
The column can be cleaned/stored following the instructions of the manufacturer and re-used for purification of the same recombinant protein at least three times.
Concentrate the eluted fraction down to 500 μL using a centrifugal concentrator (cut-off: 10 kDa) at 4 °C.
Homogenize the concentrated sample by gently pipetting up and down and transfer it to a pre-chilled 1.5 mL safe-lock Eppendorf tube. The sample is now ready to be further purified by size exclusion chromatography. It is highly recommended to proceed with size exclusion chromatography without delay, to minimize the time the protein spends in imidazole buffer.
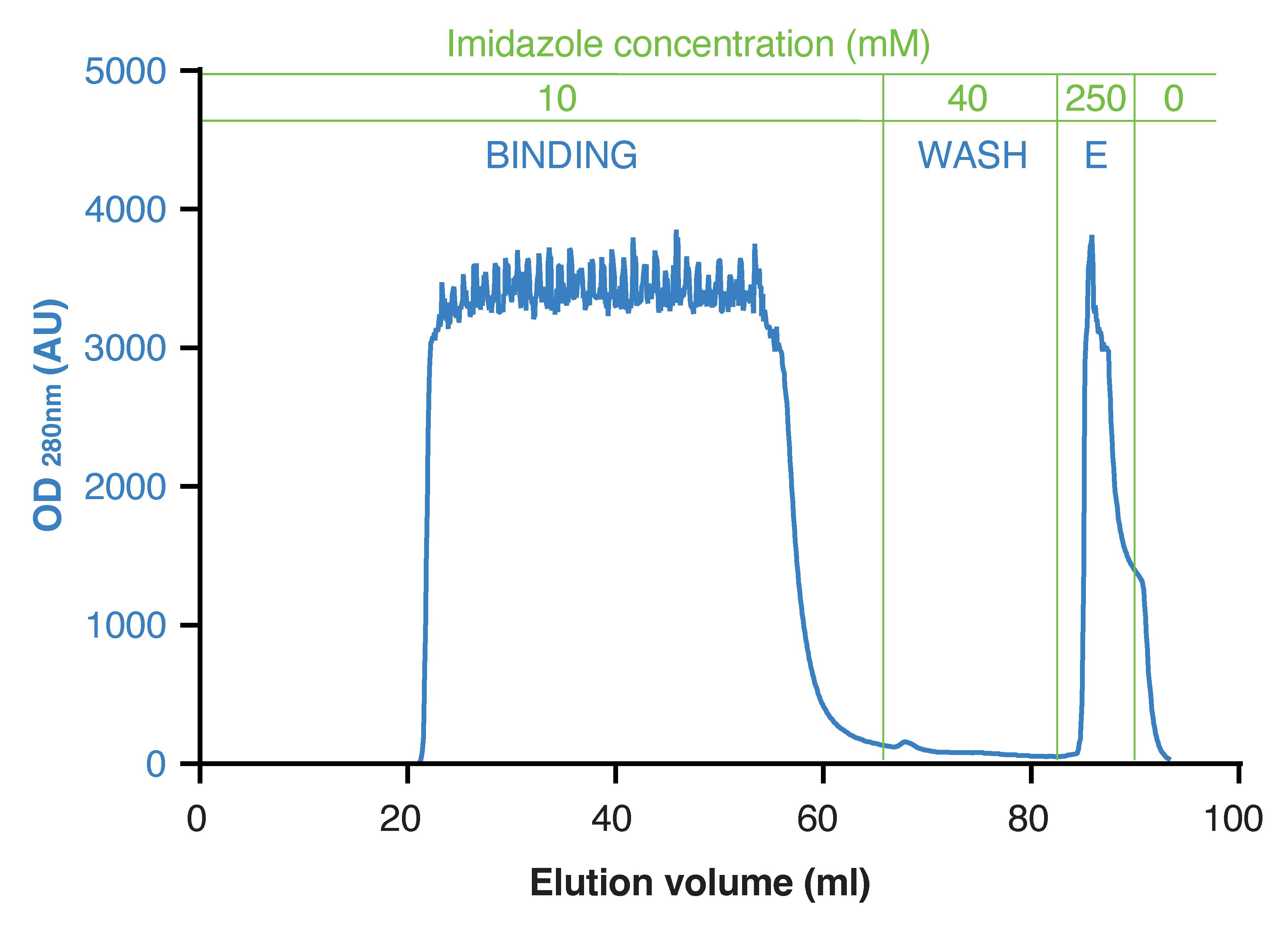
Figure 8. Elution profile resulting from immobilized metal affinity chromatography (IMAC). The adjustment of imidazole concentration in the running buffer is performed by mixing the solutions from pump A and B. OD: optical density; AU: arbitrary units; E: elution.
Purification by size exclusion chromatography (gel filtration)
Prime pump A (inlet A1) with PBS (pH 7.2).
Run the system without any column attached (flow: 0.7 mL/min) until pressure, conductivity, OD280nm, and OD215nm values are stable.
Pause the system and connect a Superdex 200 Increase 10/300 GL column to the system [set up the pressure alarm at total system pressure (~0.1 MPa) + column pressure limit (3 MPa)].
Equilibrate the column with PBS (flow: 0.7 mL/min).
Load the sample loop with 500 μL of sample to be processed. Do not pause the system.
Monitor protein elution at OD280nm and collect the fraction corresponding to the peak of interest (elution volume between 14 and 16.5 mL) (Figure 9).
The column is then cleaned/stored following the manufacturer’s instructions.
Check protein concentration.
When measurement of OD280nm is used to estimate protein concentration, the molar extinction coefficient ϵ (M-1cm-1) can be calculated from the protein amino acid sequence using the ProtParam tool (Expasy, Swiss Bioinformatics Resource Portal; https://web.expasy.org/protparam/). Protein concentration can then be calculated using the following formula: [protein]=OD280nm / (ϵ.L), where L is the distance that light travels through the solution (usually 1).
Concentrate the eluted fraction to the desired final concentration using a centrifugal concentrator (cut-off: 10 kDa) at 4 °C (check protein concentration on a regular basis during the concentration process).
Homogenize the concentrated sample, transfer it into pre-chilled 1.5 mL safe-lock Eppendorf tubes, and snap freeze in liquid nitrogen. Protein can then be stored at -80 °C.
The commonly obtained protein yield following this two-step purification process is 0.7–1 mg per liter of bacteria culture. Protein identity and purification quality can also be assessed by SDS-PAGE as previously described in this protocol (Figure 10).
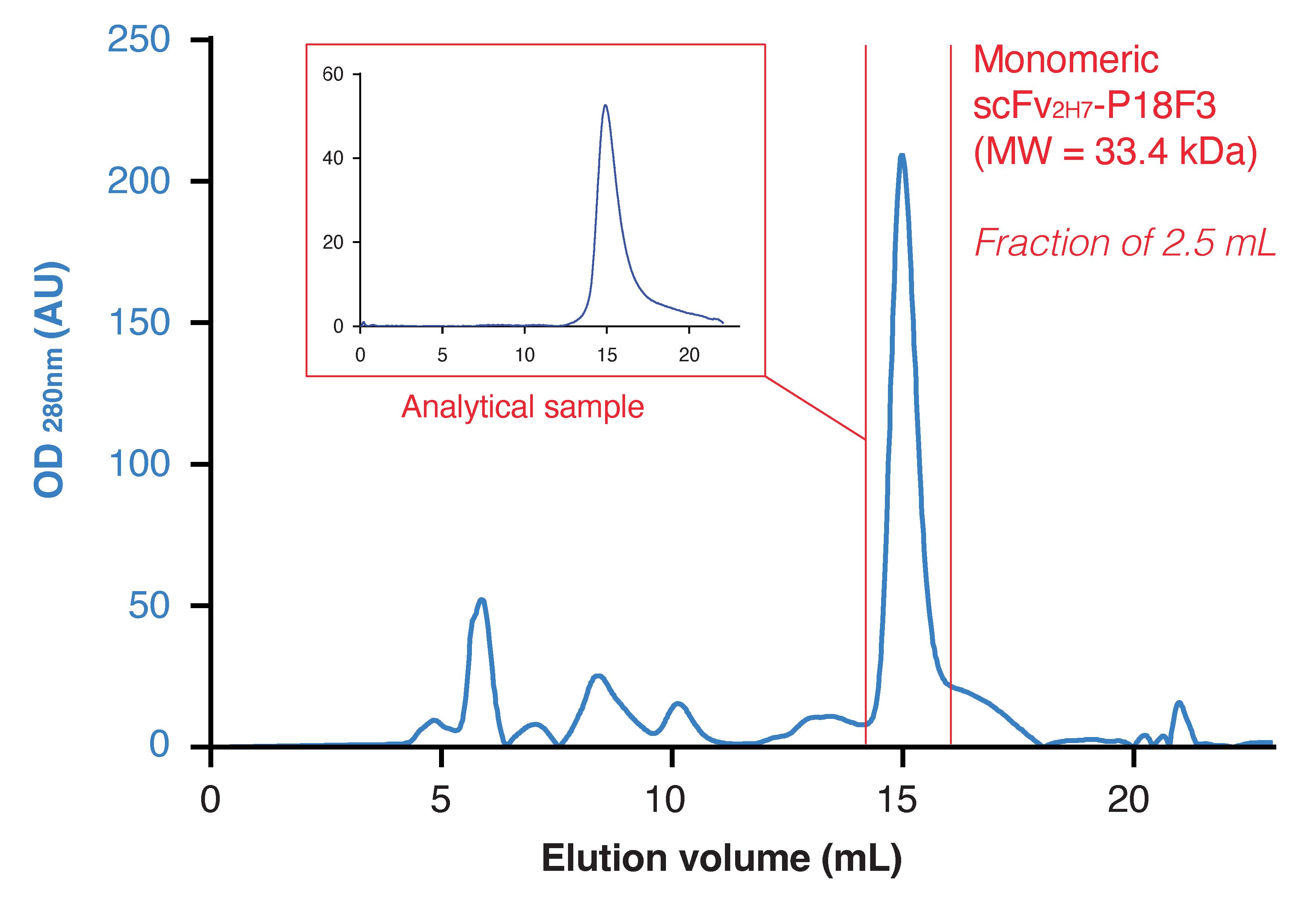
Figure 9. Elution profile resulting from size exclusion chromatography purification. Red bars delimit the peak of interest. To assess the quality of the purification, 100 μL of the eluted fraction containing scFv2H7-P18F3 was re-injected on the Superdex column (analytical sample). OD: optical density; AU: arbitrary units; E: elution.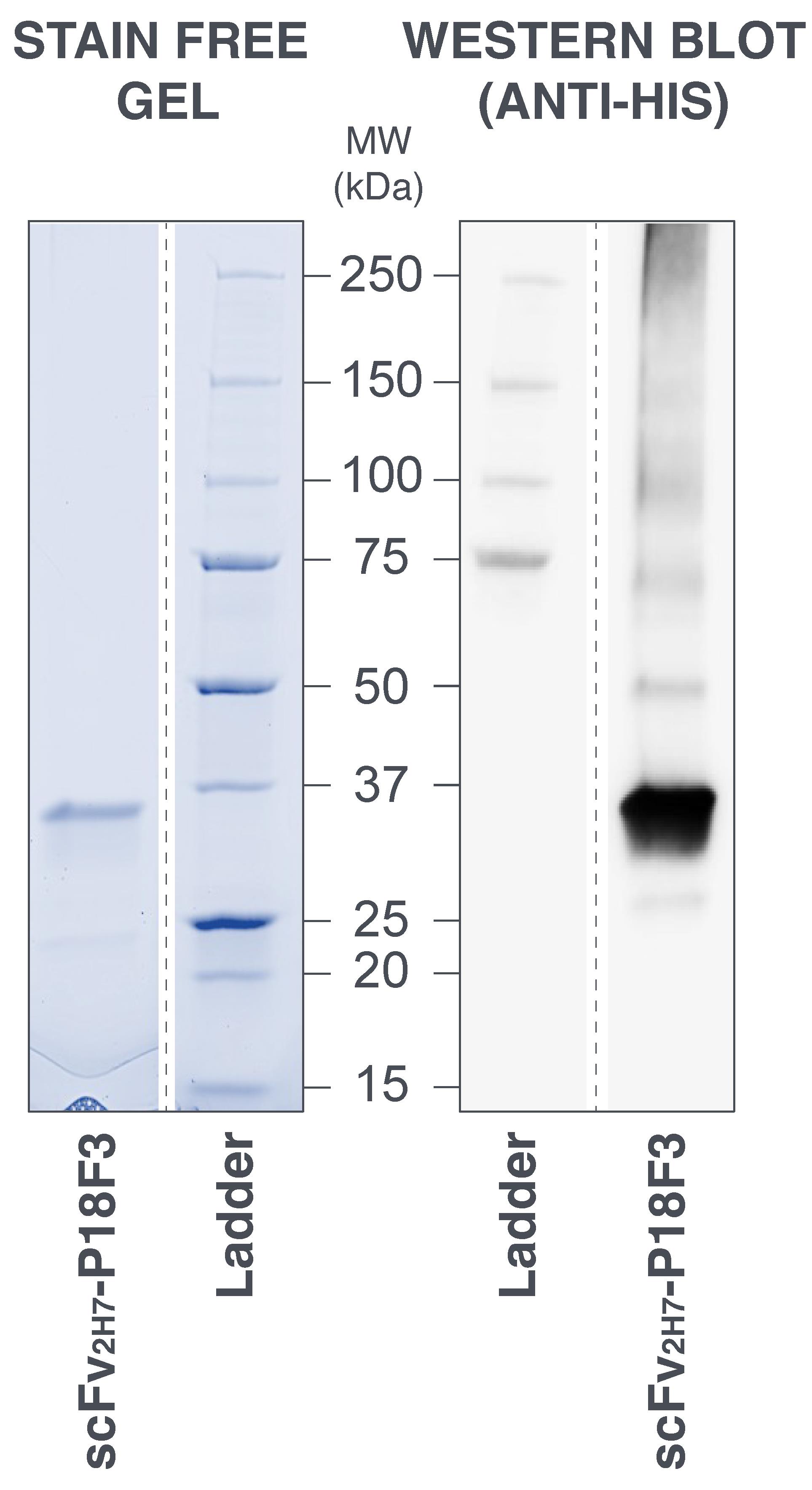
Figure 10. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of purified scFv2H7-P18F3. (Left panel) The migration profile of the eluted fraction of interest from size exclusion chromatography revealed a main band between 25 and 37 kDa with very limited amounts of visible contaminants. The gel was subjected to Stain Free protein detection (artificial colors). (Right panel) Western blot analysis performed using an anti-His antibody confirmed the identity of the purified protein. MW: molecular weight; kDa: kilo Dalton.
Recipes
LB medium (1×)
Favour the use of LB powder if possible. LB medium can alternatively be prepared according to the following recipe: 1% tryptone, 0.5% yeast extract, and 1% NaCl. Adjust to pH 7.0 and sterilize by autoclaving. Store at room temperature.
SOC medium (1×)
Favour the use of super optimal broth with catabolite repression (SOC) medium if possible. SOC medium can alternatively be prepared according to the following recipe: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose. Adjust to pH 7.0 and sterilize by autoclaving. Store at room temperature.
Kanamycin stock solution (50 mg/mL)
For 10 aliquots of 1 mL, weigh 500 mg of kanamycin sulfate powder and dissolve in 10 mL of ddH2O. Filter-sterilize using a syringe filter (0.2 μm pore size). Distribute 1 mL in ten 1.5 mL safe-lock Eppendorf tubes and freeze the aliquots at -20 °C.
50% glycerol solution
For 100 mL, mix 50 mL of sterile glycerol with 50 mL of sterile ddH20. Store at room temperature.
Bacteria resuspension solution/IMAC running buffer
50 mM Tris and 500 mM NaCl. Adjust to pH 7.2*. Store at room temperature.
IMAC elution buffer
50 mM Tris, 500 mM NaCl, and 500 mM imidazole. Adjust to pH 7.2*. Filter at 0.2 μm and degas solution. Store at room temperature.
*Note: The pH of the solution should be at least 1 pH unit lower or higher than the theoretical isoelectric point (pI) of the protein of interest to minimize protein aggregation during the purification process. The theoretical pI can be calculated from the protein amino acid sequence using the ProtParam tool (Expasy, Swiss Bioinformatics Resource Portal;https://web.expasy.org/protparam/). The theoretical pI of the BMFP scFv2H7-P18F3 is 9.23.
Tris buffer saline (TBS)
50 mM Tris-Cl and 150 mM NaCl. Adjust to pH 7.6. Store at room temperature.
Tris buffer saline tween (TBST)
50 mM Tris-Cl and 150 mM NaCl. 0.5% Tween 20. Adjust to pH 7.6. Store at room temperature.
Western blot blocking solution (TBS-5% milk)
TBS supplemented with 5 g per 100 mL of low-fat milk. Filter at 0.2 μm.
Western blot antibody dilution solution (TBST-5% milk)
TBST supplemented with 5 g/100 mL of low-fat milk. Filter using a syringe filter (0.2 μm pore size).
Phosphate buffer saline (PBS)
2.7 mM KCl, 138 mM NaCl, 1.5 mM KH2PO4, and 8 mM Na2HPO4. Adjust to pH 7.2. Filter at 0.2 μm. Store at room temperature.
Acknowledgments
This work was supported by SATT IDF-Innov, Paris, France (to B.G. and A.C.); Institut National de la Santé et de la Recherche Médicale (INSERM), France (to B.G., J.L.T. and A.C.); Institut National de la Transfusion Sanguine (INTS), Paris, France (to B.G. and A.C.).
Competing interests
BG and AC are inventors on a patent application related to this protocol filed by Institut National de la Transfusion Sanguine, Centre National de la Recherche Scientifique (CNRS), Inserm, Université de Paris (now universtité Paris Cité) (no. WO2017103020A1, filed 25 December 2015, published 22 June 2017). The authors declare that they have no other competing interests.
References
- Denoncin, K. and Collet, J. F. (2013). Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal 19(1): 63-71.
- Gamain, B., Brousse, C., Rainey, N. E., Diallo, B. K., Paquereau, C. E., Desrames, A., Ceputyte, J., Semblat, J. P., Bertrand, O., Gangnard, S., et al. (2022). BMFPs, a versatile therapeutic tool for redirecting a preexisting Epstein-Barr virus antibody response toward defined target cells. Sci Adv 8(6): eabl4363.
- Hwang, Y. C., Lu, R. M., Su, S. C., Chiang, P. Y., Ko, S. H., Ke, F. Y., Liang, K. H., Hsieh, T. Y. and Wu, H. C. (2022). Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci 29(1): 1.
- Lobstein, J., Emrich, C. A., Jeans, C., Faulkner, M., Riggs, P. and Berkmen, M. (2012). SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm.Microb Cell Fact 11: 56.
- Voge, N. V. and Alvarez, E. (2019). Monoclonal Antibodies in Multiple Sclerosis: Present and Future. Biomedicines 7(1).
- Zahavi, D. and Weiner, L. (2020). Monoclonal Antibodies in Cancer Therapy. Antibodies 9(3): 34.
Article Information
Publication history
Published: May 20, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Brousse, C., Rainey, N. E., Desrames, A., Teillaud, J. L., Gamain, B. and Chêne, A. (2023). Expression and Purification of scFv2H7-P18F3, a Bi-Modular Fusion Protein (BMFP) Targeting Human CD20. Bio-protocol 13(10): e4682. DOI: 10.21769/BioProtoc.4682.
- Gamain, B., Brousse, C., Rainey, N. E., Diallo, B. K., Paquereau, C. E., Desrames, A., Ceputyte, J., Semblat, J. P., Bertrand, O., Gangnard, S., et al. (2022). BMFPs, a versatile therapeutic tool for redirecting a preexisting Epstein-Barr virus antibody response toward defined target cells. Sci Adv 8(6): eabl4363.
Category
Immunology
Cancer Biology > Tumor immunology > Cancer therapy
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







