- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Optimized Protocol for Detecting Guard Cell–specific Gene Expression by in situ RT-PCR in Brassica rapa
(*contributed equally to this work) Published: Vol 13, Iss 17, Sep 5, 2023 DOI: 10.21769/BioProtoc.4810 Views: 272
Reviewed by: Alessandro DidonnaSailendra SinghAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
An In-depth Guide to the Ultrastructural Expansion Microscopy (U-ExM) of Chlamydomonas reinhardtii
Nikolai Klena [...] Virginie Hamel
Sep 5, 2023 1042 Views

Absolute Quantification of mRNA Isoforms in Adult Stem Cells Using Microfluidic Digital PCR
Shubhangi Das Barman [...] Antoine de Morree
Sep 5, 2023 260 Views
Fluorescence in situ Localization of Pri-miRNAs in Isolated Arabidopsis thaliana Nuclei
Tomasz Gulanicz [...] Artur Jarmolowski
Sep 20, 2023 364 Views
Abstract
Since the genetic transformation of Chinese cabbage (Brassica rapa) has not been well developed, in situ RT-PCR is a valuable option for detecting guard cell–specific genes. We reported an optimized protocol of in situ RT-PCR by using a FAMA homologous gene Bra001929 in Brassica rapa. FAMA in Arabidopsis has been verified to be especially expressed in guard cells. We designed specific RT-PCR primers and optimized the protocol in terms of the (a) reverse transcription time, (b) blocking time, (c) antigen-antibody incubation time, and (d) washing temperature. Our approach provides a sensitive and effective in situ RT-PCR method that can detect low-abundance transcripts in cells by elevating their levels by RT-PCR in the guard cells in Brassica rapa.
Keywords: Brassica rapaBackground
In situ RT-PCR combines RT-PCR and in situ hybridization technologies to amplify specific nucleic acid sequences in cells or tissue sections, enabling the localization of specific low-copy number sequences to be detected by immunohistochemistry (Nuovo, 2001). This technique identifies mRNA in the tissues; it was initially used to locate and detect viral gene expression in human and animal cells (Martinez, 1995; Bates et al., 1997; Hoyland et al., 1997; Kher and Bacallao, 2001; Cubas-Nuñez et al., 2017) and was then introduced into plant research. Thus far, this technology has been successfully applied in many plants including pea, Arabidopsis (Deeken and Kaldenhoff, 1997), cucumber (Urbańczyk-Wochniak et al., 2002), tomato (Portillo et al., 2013), barley (Ferdous et al., 2017), and barley and Arabidopsis (Athman et al., 2014); the protocol from Athman et al. (2014) has been widely used in plant research (Hocking et al., 2017; Olsen and Krause, 2019). Bra001929 is a homolog in Brassica rapa to the guard cell–specific gene FAMA in Arabidopsis, which has been identified and analyzed in a wide range of plant species. The Bra001929 gene can also be used as a marker gene for Chinese cabbage guard cells.
Materials and reagents
Leaf epidermis
Double-edged blade (Gillette)
RNaseZap® solution (Ambion, catalog number: AM9786)
Formalin (Sigma, catalog number: F8775-25ML)
Acetic acid (Sigma, catalog number: 695092-500ML)
Ethanol absolute (Sangon Biotech, catalog number: A500737-0500)
DEPC (Sigma, catalog number: D5758-25ML)
Nuclease-free 10× PBS (Solarbio, catalog number: P1022)
RNaseOUTTM recombinant ribonuclease inhibitor (Invitrogen, catalog number: 10777-019)
10× TURBO DNaseTM buffer (Invitrogen, TURBO DNAfreeTM kit, catalog number: AM1907)
DNase I, RNase-free (Invitrogen, catalog number: EN0521)
0.5 M EDTA, pH 8.0, RNase-free (Ambion, catalog number: AM9260G)
EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, catalog number: AE311-02)
DIG-11-dUTP (Roche, catalog number: 11093088910)
Bovine serum albumin (BSA) (Sigma, catalog number: A6003-100G)
BM Purple AP Substrate (Roche, catalog number: 11442074001)
Tris (Biotopped, catalog number: T6061)
NaCl (Solarbio, catalog number: S8210)
Glycerol (Sigma, catalog number: 56-81-5)
Dilute Anti-DIG-AP in 1% BSA blocking solution (see Recipes)
DNase/RNase free water (see Recipes)
FAA fixative (see Recipes)
Washing buffer I (see Recipes)
1× PBS (see Recipes)
1% BSA blocking solution (see Recipes)
40% glycerol solution (see Recipes)
Equipment
Pointed tweezers (AsOne, catalog number: CC-9820-01)
Super frost plus slides, 76 mm × 25 mm (Fisher Scientific, catalog number: 12-550-15)
Cover glass (CITOGLAS, catalog number: BZ1164)
Frame-SealTM incubation chambers (Bio-Rad, catalog number: SLF-0601)
15 mL centrifuge tube (Corning, catalog number: 433052)
2 mL centrifuge tube (Axygen, catalog number: MCT-060-A)
0.2 mL centrifuge tube (Axygen, catalog number: PCR-02-C)
Metal constant temperature incubator (Coyotebio, catalog number: H2O3-100C)
Water bath thermal cycler, an automated high throughput water bath thermal cycler with flexible modular design for rapid temperature change and precise control. Key technical parameters: three tanks, temperature controlled 40–97 °C (HC Scientific, catalog number: ID 156 B03)
Optical microscope (Zeiss, catalog number: 3150012307)
Software
ImageJ (https://imagej.net/software/imagej/)
Microsoft Excel (Microsoft Corp., Albuquerque, NM, USA)
Procedure
Tissue fixation
First make 20 mL of FAA fixative (see Recipes): 2% formalin, 5% acetic acid, and 60% ethanol well mixed; place in refrigerator at 4 °C.
Fix the Frame-SealTM incubation chambers on the glass slides and add 150 μL of pre-cooled FAA fixative to each glass slide.
Rinse the blade with sterile water before tissue fixation, quickly tear off the epidermis with pointed tweezers, and immediately place it in an ice bath in FAA fixative.
Place the glass slide under the optical microscope to detect whether the epidermal tissue is stuck with mesophyll tissue. If so, then peel the epidermis again; if not, proceed to the next step.
The sample needs to be fixed 3–4 times, 15 min each. Try to remove it with a pipette every time the fixative is changed. During fixation, make washing buffer 1:5% acetic acid and 60% ethanol, mix well, and place in the refrigerator at 4 °C.
After fixation, add 150 μL of washing buffer 1 to each slide to wash the tissue samples on ice three times for 8–10 min each time. To ensure sufficient washing, the slide can be rotated slightly.
Add 150 μL of 1× PBS to each slide and continue washing three times for 3 min each on ice to maintain the ion concentration; finally, add the premix to each slide: 102 μL of RNase-free water + 3 μL of RNaseOUTTM ribonuclease (RNase) inhibitor (see Figure 1).
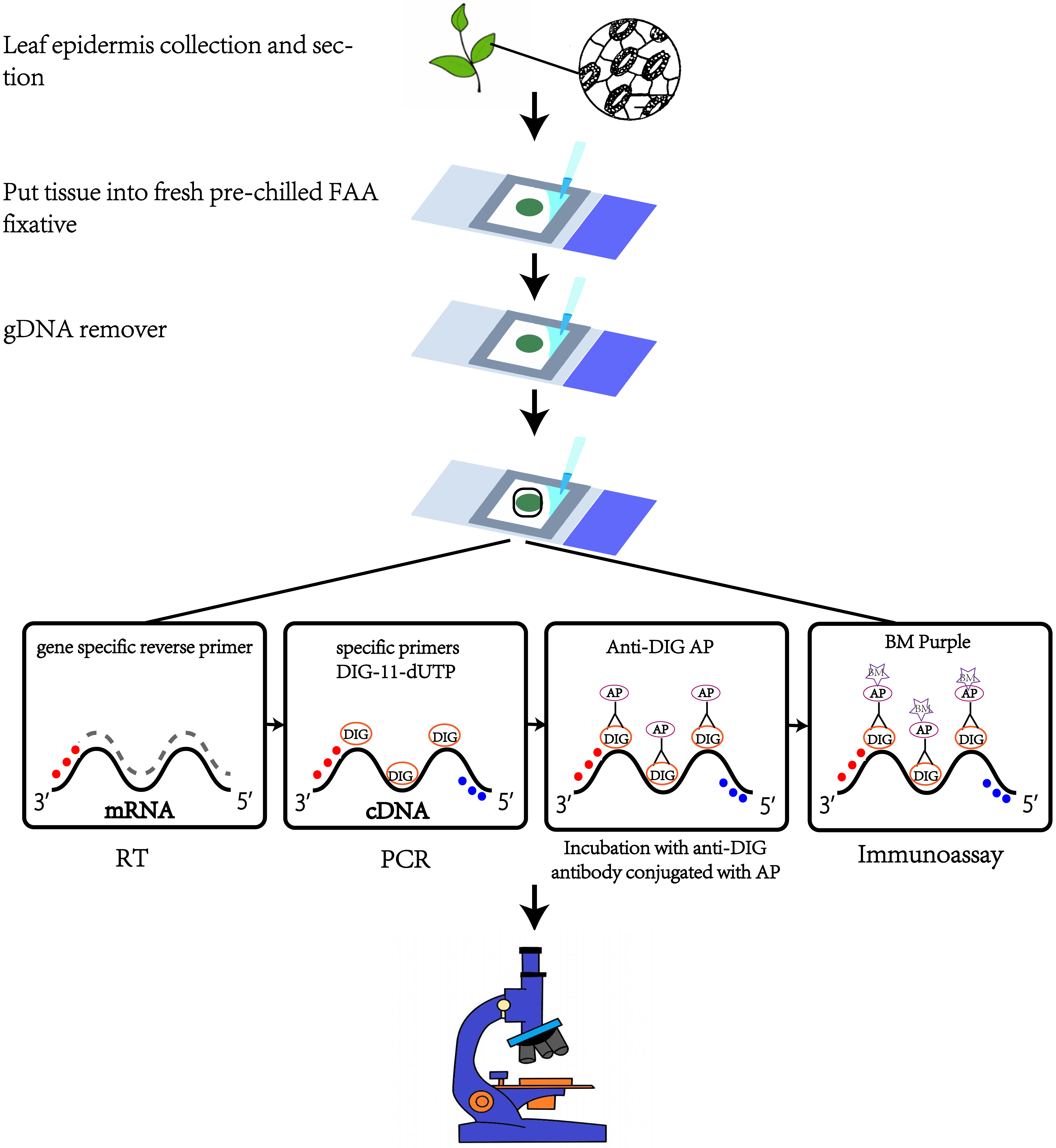
Figure 1. Schematic of in situ RT-PCR procedure
Removal of genomic DNA
Configuration of master mix: 12 μL of 10× TURBO DNaseTM buffer + 3 μL of 1UμL -1 DNase I. After mixing, add to the sample in a final volume of 120 μL and cover with a cover glass.
Adjust the temperature of the metal bath to 37 °C in advance and place the glass slide in the metal bath for 1 h.
Adjust the temperature of the metal bath to 70 °C, carefully remove the coverslip with tweezers, add 4 μL of 0.5 M EDTA pH 8.0 to the slide, cover the coverslip to inactivate for 15 min, and store at 4 °C in the refrigerator for approximately 5 min.
Use tweezers to carefully remove the cover glass. Since the epidermis may stick to the cover glass during high-temperature incubation, you can use a sterile scalpel to move it into the solution and a pipette to remove as much DNase solution as possible.
Pipette 200 μL of pre-cooled DEPC water into the slide to wash the sample on ice. To ensure sufficient washing, the slide can be rotated slightly.
In order not to affect subsequent experiments, discard as much RNase-free water as possible with a pipette after washing (see Figure 1).
Reverse transcription
In order to obtain higher synthesis efficiency, first configure the primer and RNase-free water master mix: 3 μL of 10 μM gene-specific reverse primer + 84.6 μL of RNase-free water.
Immediately add the premix to the glass slide that has removed RNase-free water and cover it with a cover glass.
Adjust the temperature of the metal bath to 65 °C in advance, incubate the slides at 65 °C for 8 min, and prepare the RT reaction solution during the incubation.
After the incubation, adjust the temperature of the metal bath to 42 °C; then, use the pointed tweezers to carefully remove the cover glass and add the RT premix (Table 1) after the slide is ice-bathed for 2 min. To avoid false positives during the in situ RT-PCR process, we used the control in which the reverse transcription enzyme was not used in the RT reaction.
Table 1. RT reaction premix
Component Volume 2× ES Reaction Mix 30 μL EasyScript® RT Enzyme Mix 1.2 μL RNaseOUTTM ribonuclease (RNase) inhibitor 1.2 μL Bubbles may be present after adding the RT premix; use a 10 μL pipette to carefully pipette them out and then cover the cover glass.
Place the slides in a metal bath and incubate at 42 °C for 1.5 h. Then, adjust the temperature of the metal bath to 85 °C and incubate for 1 min to inactivate the reverse transcriptase. After incubation, remove the slides and place in the refrigerator at 4 °C for 5 min (see Figure 1).
In situ PCR
Because the temperature of the water bath PCR thermal cycler is low, it is necessary to turn on the thermal cycler to preheat at the beginning of the experiment. The PCR reaction is premixed (Table 2). Configure the in situ PCR mix when the slide is placed at 4 °C.
Table 2. Primer design
Primer Primer sequence Bra001929F 5′-GAA CAA GTC GTG CTT GGC TG-3′ Bra001929R 5′-CAC GTG TTT ATA CCT ACT TGC CT-3′ Vortex and briefly centrifuge the configured in situ PCR mix and place it in a refrigerator at 4 °C.
Use tweezers to carefully remove the cover glass and remove the RT reaction solution.
Take 200 μL of pre-cooled RNase-free water to wash the tissue three times on ice. To ensure sufficient washing, the slide can be rotated slightly.
In order not to affect the subsequent PCR, use a pipette to discard as much RNase-free water as possible after washing.
Add 90 μL of pre-cooled in situ PCR mix (in Table 3) to each sample. If there are bubbles, use a 10 μL pipette to carefully suck them out. Finally, carefully glue the polyester covers and press them firmly along the edge of the frame. Avoid squeezing out the PCR mix.
The primers for Bra001929 were designed according to the following guidelines: primer length of 18–24 bp, GC content of 40%–60%, with the last five bases of the 3' end being less rich in GC, which resulted in the pairs of primers for Bra001929.
The suggested PCR cycle setting is in Table 4. However, annealing temperature and extension time are adjusted according to primer and product size (see Figure 1).
Table 3. PCR reaction
Component Volume H2O 64.94 μL 5× Phusion HF Buffer* 18 μL 10 mM dNTPs 1.8 μL Forward primer 4.5 μL Reverse primer 4.5 μL Phusion DNA Polymerase 0.9 μL DIG-11-dUTP 0.36 μL Table 4. PCR cycling instructions
Cycle step 3-step protocol Cycles Temperature Time Initial denaturation 98 °C 30 s 1 Denaturation 98 °C 10 s 35 Denaturation 55 °C 25 s Extension 72 °C 5 s Final extension 72 °C 7 min 1
Immunoassay
The frame and the polyester covers are very tightly bonded. If you tear it directly, the frame will also be torn off the glass slide. So Incubate the slide on ice and use a sterile scalpel to cut off the polyester covers along the frame.
After cutting off the polyester covers, carefully remove the PCR mix.
Pipette 200 μL of pre-cooled 1× PBS to wash the tissue on ice three times for 3 min each. To ensure sufficient washing, the slide can be rotated slightly.
Remove as much 1× PBS as possible after each wash.
Adjust the temperature of the metal bath to 37 °C in advance; add 100 μL of 1% BSA blocking solution to the tissue sample, rotate the slide slightly to mix, and incubate at room temperature for 30 min.
Dilute the alkaline phosphatase–labeled anti-Digoxigenin antibody at 1:500 in fresh 1% BSA blocking solution. Each sample needs to be configured with 80 μL: 0.16 μL of Anti-Digoxigenin-AP + 79.84 μL of 1% BSA blocking solution. Store in the refrigerator at 4 °C.
After blocking at room temperature, carefully remove the 1% BSA blocking solution and pipette 100 μL of alkaline phosphatase–labeled anti-digoxigenin antibody binding solution to the tissue sample and incubate at room temperature for 1 h.
After the antigen and antibody incubation, carefully remove the antibody binding solution.
Add 150 μL of elution buffer II to carefully rinse the tissue sample three times, each for 10 min.
After elution, add 70 μL of alkaline phosphatase substrate BM Purple to the tissue sample in the dark (turn the alkaline phosphatase substrate upside down and mix well before use) and leave it at room temperature for 30 min without a cover glass. Then, observe under the optical microscope every 15 min (10× magnification).
Once the purple-blue amplification signal is detected, find the best place to take pictures and archive. Then, remove the alkaline phosphatase substrate, pipette 150 μL of elution buffer II into the slide to wash the sample on ice three times for 2 min each, and then wash once with sterile water on ice. Finally, add 40 μL of 40% glycerol for mounting and cover with a coverslip (see Figure 1).
Data analysis
The positive guard cells exhibited significant hybridization signals using the optimized protocol based on three duplicates (Figure 2). Here, we used the optical density value to assess the hybridization signal intensity. First, ImageJ (1.53c; Wayne Rasband software; National Institutes of Health; USA) was used for image analysis to obtain the optical density of the region of interest (ROI) and the Area (Area of the selected range) and IntDen (IOD of the selected range) of positive guard cells, positive epidermal cells, negative guard cells, and negative epidermal cells in the results. The ratio of IntDen to Area is the average optical density value of a single cell. Data were analyzed using Microsoft Excel (Microsoft Corp., Albuquerque, NM, USA). The results were expressed as mean ± S.E.M. (standard error of the mean, see Figure 3).
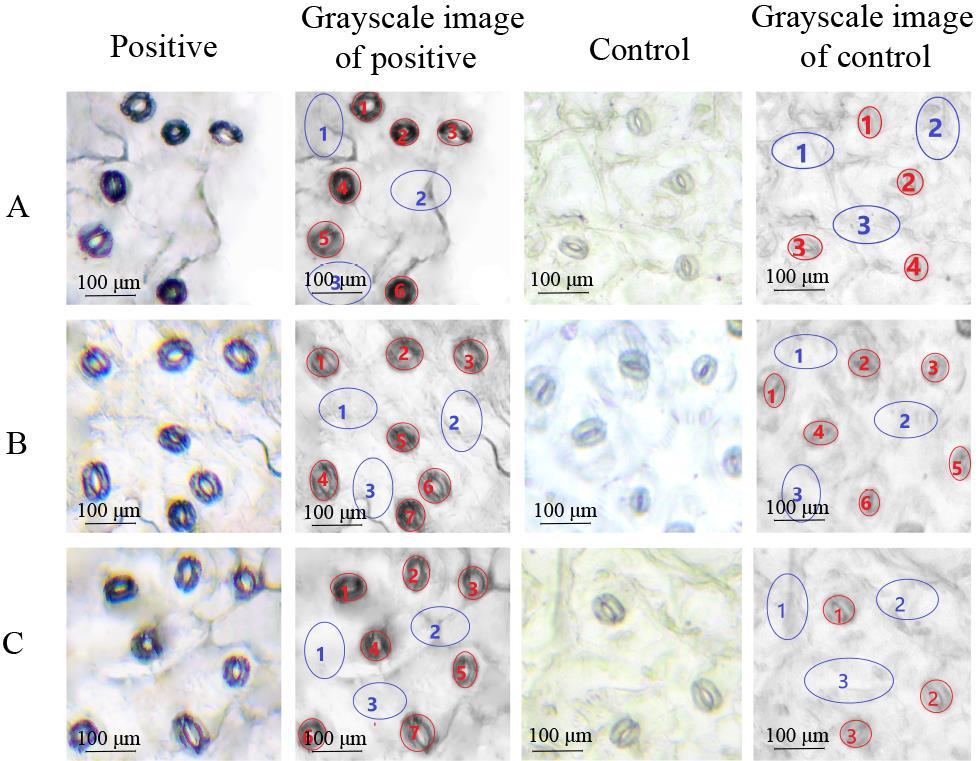
Figure 2. Three duplicates of in situ RT-PCR in B. rapa guard cells. A. First duplicate with visualization of the transcripts of the target genes in the guard cells. B. Second duplicate. C. Third duplicate. Red letters represent guard cells and blue letters represent epidermal cells. The control without RT showed no visualization of transcripts in the guard cells. The 8-bit grayscale images were converted by ImageJ software. Scale bars represent 100 μm.
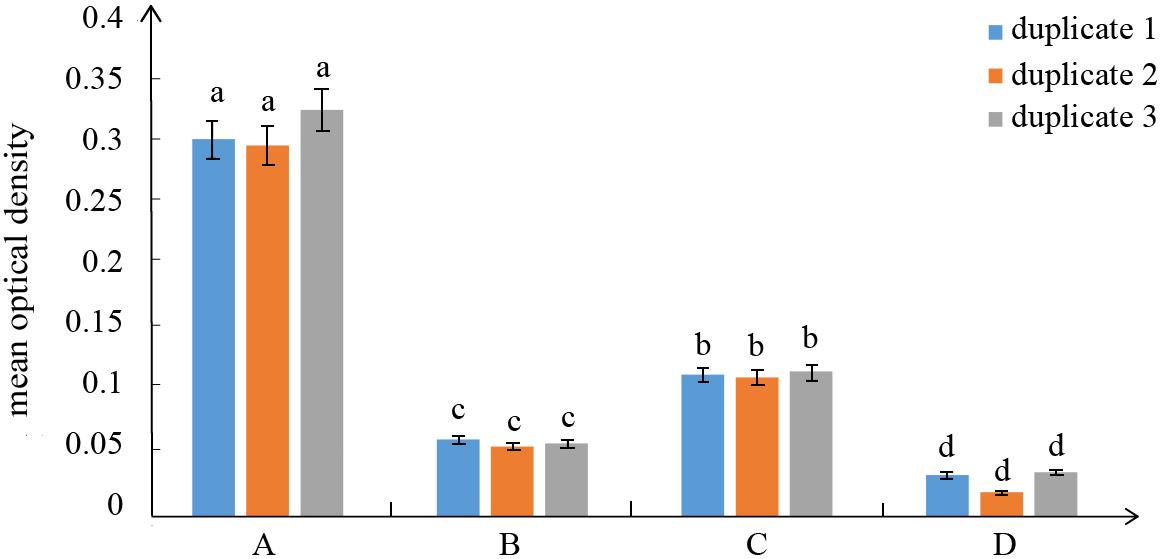
Figure 3. Mean optical density values of three duplicates for different cells in the in situ RT-PCR. A. Mean optical density values of the positive guard cells correspond to the grayscale image of the positive cells marked in red letters (Figure 2b, 2f, 2j). B. Mean optical density values of the control epidermal cells in the positive slides correspond to the grayscale image of the positive cells marked with blue letters (Figure 2b, 2f, 2j). C. Mean optical density values of the control guard cells correspond to the grayscale image of the control cells marked with red letters (Figure 2d, 2h, 2l). D. Mean optical density values of the control epidermal cells correspond to the grayscale image of the control cells marked with blue letters (Figure 2d, 2h, 2l). The error was calculated from the standard deviation of the cell average and different letters on top of the bars indicate significant differences.
Recipes
DNase/RNase free water
Add 1 mL of DEPC to 1,000 mL of deionized water to prepare 0.1% DEPC water. Put it in a 1,000 mL blue cap bottle, shake and mix thoroughly, let it stand for 24 h, and then high pressure for 40 min to remove DEPC.
FAA fixative
2% formalin, 5% acetic acid, 60% ethanol
Washing buffer I
5% acetic acid, 60% ethanol
DNase/RNase free water
Add 1 mL of DEPC to 1,000 mL of deionized water to prepare 0.1% DEPC water. Put it in a 1,000 mL blue cap bottle, shake and mix thoroughly, let it stand for 24 h and then high pressure for 40 min to remove DEPC.
1× PBS
1 mL of 10× PBS + 9 mL of H2O
1% BSA blocking solution
Dilute Anti-DIG-AP in 1% BSA blocking solution: 0.16 μL of Anti-Digoxigenin-AP + 79.84 μL of 1% BSA blocking solution.
Washing buffer I
100 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl
40% glycerol solution
4 mL of glycerol solution + 6 mL of H2O
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Grant No. 31630068), the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, China. This protocol was modified based on the previous work from Athman et al. (2014).
Competing interests
The authors declare no conflicts of interest.
References
- Athman, A., Tanz, S. K., Conn, V. M., Jordans, C., Mayo, G. M., Ng, W. W., Burton, R. A., Conn, S. J. and Gilliham, M. (2014). Protocol: a fast and simple in situ PCR method for localising gene expression in plant tissue. Plant Methods 10: 29.
- Bates, P. J., Sanderson, G., Holgate, S. T. and Johnston, S. L. (1997). A comparison of RT-PCR, in-situ hybridisation and in-situ RT-PCR for the detection of rhinovirus infection in paraffin sections. J. Virol. Methods 67(2): 153–160.
- Ferdous, J., Sanchez-Ferrero, J. C., Langridge, P., Milne, L., Chowdhury, J., Brien, C. and Tricker, P. J. (2017). Differential expression of microRNAs and potential targets under drought stress in barley. Plant Cell Environ. 40(1): 11–24.
- Hoyland, J. A., Mee, A. P., Baird, P., Braidman, I. P., Mawer, E. B. and Freemont, A. J. (1997). Demonstration of estrogen receptor mRNA in bone using in situ reverse-transcriptase polymerase chain reaction. Bone 20(2): 87–92.
- Hocking, B., Conn, S. J., Manohar, M., Xu, B., Athman, A., Stancombe, M. A., Webb, A. R., Hirschi, K. D. and Gilliham, M. (2017). Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J. Exp. Bot. 68(15): 4171–4183.
- Kher, R. and Bacallao, R. (2001). Direct in situ reverse transcriptase-polymerase chain reaction. Am. J. Physiol. Cell Physiol. 281(2): C726–732.
- Deeken, R. and Kaldenhoff, R. (1997). Light-repressible receptor protein kinase: a novel photo-regulated gene from Arabidopsis thaliana. Planta 202(4): 479–486.
- Cubas-Nuñez, L., Duran-Moreno, M., Castillo-Villalba, J., Fuentes-Maestre, J., Casanova, B., Garcia-Verdugo, J. M. and Gil-Perotin, S. (2017). In situ RT-PCR Optimized for Electron Microscopy Allows Description of Subcellular Morphology of Target mRNA-Expressing Cells in the Brain. Front. Cell Neurosci. 11: 141.
- Martinez, A., Miller, M. J., Quinn, K., Unsworth, E. J., Ebina, M. and Cuttitta, F. (1995). Non-radioactive localization of nucleic acids by direct in situ PCR and in situ RT-PCR in paraffin-embedded sections. J. Histochem. Cytochem. 43(8): 739–747.
- Portillo, M., Cabrera, J., Lindsey, K., Topping, J., Andres, M. F., Emiliozzi, M., Oliveros, J. C., Garcia-Casado, G., Solano, R., Koltai, H., et al. (2013). Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197(4): 1276–1290.
- Nuovo, G. J. (2001). Co-labeling using in situ pcr: A review. J. Histochem. Cytochem. 49(11): 1329–1339.
- Olsen, S. and Krause, K. (2019). A rapid preparation procedure for laser microdissection-mediated harvest of plant tissues for gene expression analysis. Plant Methods 15: 88.
- Urbańczyk-Wochniak, E., Filipecki, M. and Przybecki, Z. (2002). A useful protocol for in situ RT-PCR on plant tissues. Cell Mol. Biol. Lett. 7(1): 7–18.
Article Information
Publication history
Published: Sep 5, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Song, Y., Guo, X., Wu, J., Liang, J., Lin, R., Yan, Z. and Wang, X. (2023). An Optimized Protocol for Detecting Guard Cell–specific Gene Expression by in situ RT-PCR in Brassica rapa. Bio-protocol 13(17): e4810. DOI: 10.21769/BioProtoc.4810.
Category
Plant Science > Plant cell biology > Cell imaging
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







