- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An in vitro Assay to Probe the Formation of Biomolecular Condensates
Published: Vol 13, Iss 17, Sep 5, 2023 DOI: 10.21769/BioProtoc.4813 Views: 759
Reviewed by: Emilia KrypotouLu LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Characterising Plant Deubiquitinases with in vitro Activity-based Labelling and Ubiquitin Chain Disassembly Assays
Michael J. Skelly and Steven H. Spoel
May 5, 2021 3482 Views
High Throughput Analyses of Ascorbate-turnover Enzyme Activities in Rice (Oryza sativa L.) Seedlings
Lin-Bo Wu [...] Michael Frei
Oct 20, 2021 2130 Views
An in vitro Coupled Assay for PEPC with Control of Bicarbonate Concentration
Nicholas R. Moody [...] James D. Reid
Dec 20, 2021 1438 Views
Abstract
Biomolecular condensates are membrane-less assemblies of proteins and nucleic acids formed through liquid–liquid phase separation (LLPS). These assemblies are known to temporally and spatially regulate numerous biological activities and cellular processes in plants and animals. In vitro phase separation assay using recombinant proteins represents one of the standard ways to examine the properties of proteins undergoing LLPS. Here, we present a detailed protocol to investigate in vitro LLPS using in vitro expressed and purified recombinant proteins.
Keywords: Biomolecular condensatesBackground
Liquid–liquid phase separation (LLPS) is mostly triggered by multivalent interactions between macromolecules including proteins and nucleic acids (Boeynaems et al., 2018). Several factors, such as protein concentration, pH, post-translational modifications, or the presence of other molecules, affect the process of phase separation by affecting the multivalent interactions (Saha et al., 2016; Dao et al., 2018; Dignon et al., 2020; Li et al., 2022). Both in vitro and in vivo strategies have been developed to test whether a protein undergoes LLPS, including the in vitro phase separation using recombinant proteins and fluorescence recovery after photobleaching (FRAP). In vitro phase separation assay offers a simple and fast detection method and is necessary to determine whether a protein undergoes phase separation. This approach could be used to evaluate any protein of interest. In this protocol, we take GFP and GFP-HRLP (Zhang et al., 2022) as examples to present a step-by-step guide for this approach.
Materials and reagents
pGEX-6p-2 vector (Pharmacia, 27-4598-01)
Primers used to construct GST-GFP and GST-GFP-HRLP:
GST-GFP-F-BamHI: 5′-CGCGGATCCATGAGTAAAGGAGAAGAACT-3′
GST-GFP-R-EcoRI: 5′-CCGGAATTCTTTGTATAGTTCATCCATGC-3′
HRLP-F-SalI: 5′-ACGCGTCGACTCATGCCACCGAAGGTTGTGAAG-3′
HRLP-R-NotI: 5′-AAGGAAAAAAGCGGCCGCTCAGTAGTATGATCCTGGAC-3′
Rosetta (DE3) competent cells (Novagen, catalog number: 70954)
Ampicillin (Sigma-Aldrich, CAS number: 7177-48-2)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, CAS number: 367-93-1)
Glutathione Sepharose beads (GE Healthcare, catalog number: GE17-0756-05)
PreScission protease (GE Healthcare, catalog number: 27-0843-01)
Bio-Rad protein assay dye reagent concentrate (Bio-Rad, catalog number: 5000006)
Bovine serum albumin (BSA) (Sigma-Aldrich, CAS number: 9048-46-8)
Precision Plus protein standard all blue marker (Bio-Rad, catalog number: 1610373)
PEG-8000 (Sigma-Aldrich, catalog number: 25322-68-3)
Hole microscope slide (BoliOptics, catalog number: SL39101004)
Cover glass (TRUSCO, catalog number: 122-9777)
Tryptone (Thermo Fisher Scientific, catalog number: 211705)
NaCl (Sigma-Aldrich, CAS number: 7647-14-5)
Yeast extract (Thermo Fisher Scientific, catalog number: 211929)
Agar (for LB plates) (BD, catalog number: 214010)
Methanol (Sigma-Aldrich, CAS number: 67-56-1)
Acetic acid (Sigma-Aldrich, CAS number: 64-19-7)
Coomassie Brilliant Blue R-250 (Bio-Rad, catalog number: 1610400)
Tris (Vivantis, catalog number: PR0612)
EDTA (Sigma-Aldrich, CAS number: 60-00-4)
Triton X-100 (Bio-Rad, catalog number: 1610407)
DTT (Roche, CAS number: 3483-12-3)
Complete EDTA-free protease inhibitor cocktail (Roche, catalog number: 5056489001)
Glycine (Bio-Rad, catalog number: 1610718)
SDS (Vivantis, catalog number: 151-21-3)
Acrylamide/bis 37.5:1 (Bio-Rad, catalog number: 1610158)
Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: 7727-54-0)
TEMED (Bio-Rad, catalog number: 1610801)
LB medium (see Recipes)
Coomassie blue staining buffer (see Recipes)
Destaining solution (see Recipes)
Lysis buffer (see Recipes)
Cleavage buffer (see Recipes)
SDS running buffer (see Recipes)
10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (see Recipes)
Recipes
LB medium
10 g/L tryptone
10 g/L NaCl
5 g/L yeast extract
15 g/L agar (for LB plates)
Coomassie blue staining buffer
20% methanol
10% acetic acid
0.1% Coomassie Brilliant Blue R-250
Destaining solution
20% methanol
10% acetic acid
Lysis buffer
10 mM Tris-HCl pH 8.0
15 mM NaCl
1 mM EDTA
1% Triton X-100
5 mM DTT
1× Complete EDTA-free protease inhibitor cocktail
Cleavage buffer
50 mM Tris-HCl pH 7.0
150 mM NaCl
1 mM EDTA
1 mM DTT
0.01% Triton X-100
SDS running buffer
3.03 g/L Tris
14.4 g/L glycine
1 g/L SDS
10% SDS-PAGE gel (for two 1.0 mm gels)
Separation gel:
4.1 mL of H2O
3.3 mL of 30% acrylamide/bis 37.5:1
2.5 mL of 1.5 M Tris-HCl pH 8.8
100 μL of 10% SDS
100 μL of APS
10 μL of TEMED
Stacking gel:
3.05 mL of H2O
0.65 mL of 30% Acrylamide/bis 37.5:1
0.625 mL of 1 M Tris-HCl pH 6.8
50 μL of 10% SDS
50 μL of APS
5 μL of TEMED
Equipment
Incubator (Panasonic, catalog number: 24127-88)
Shaker (INFORS, model: SKU BLE2000100)
Sonicator (Sonics, model: VCX-130)
Microcentrifuge (Thermo Fisher Scientific, catalog number: 75003424)
Centrifuge (Beckman Coulter, model: Allegra X-22)
Nutating mixer (Labnet, model: S0500-230V-EU)
Protein vertical electrophoresis cell (Bio-Rad, catalog number: 658004)
PowerPac power supply (Bio-Rad, catalog number: 1645070)
Confocal microscope (Olympus, model: FV3000)
Software
FV31-SW (Olympus)
Procedure
Recombinant protein expression
Clone the coding sequences of GFP and GFP fused with HRLP into the pGEX-6p-2 vector to generate GST-GFP and GST-GFP-HRLP constructs, respectively.
Transform the constructs of GST-GFP and GST-GFP-HRLP into DE3 competent cells.
Spread the transformed competent cells onto LB plates containing 100 mg/L ampicillin and incubate overnight at 37 °C to form colonies.
Inoculate one single colony into 2 mL of LB liquid medium supplemented with ampicillin at the final concentration of 100 mg/L and incubate at 37 °C overnight with vigorous shaking at 220 rpm.
Transfer the overnight cell culture to 200 mL of LB liquid medium supplemented with 100 mg/L ampicillin and grow at 37 °C with shaking at 220 rpm until the OD600 value reaches 0.6–1.0.
Add IPTG at the final concentration of 0.2 mM to the cultured cells to induce proteins of interest at 16 °C overnight (Note 1).
Test the solubility of GST-GFP and GST-GFP-HRLP fusion proteins by running a protein gel followed by Coomassie blue staining through the following steps:
Transfer 2 mL of induced and non-induced cell cultures to Eppendorf tubes, followed by centrifugation at maximum speed for 1 min to pellet the cells. Meanwhile, keep the rest of the cell cultures at 4 °C.
Add 1 mL of lysis buffer (Note 2) to resuspend the cells and further break cells by sonication (output watt at 6, 30 s on/30 s off) until the solution becomes clear.
Centrifuge the lysed cells at maximum speed at 4 °C for 10 min and transfer the supernatant into a new 1.5 mL tube as the soluble proteins.
Add 6× SDS loading dye to the supernatant and denature the proteins at 100 °C for 10 min.
Run an SDS-PAGE gel. After running is completed, stain the gel with Coomassie blue staining buffer for at least 1 h at room temperature.
Destain the gel by soaking the gel in destaining solution. Induced recombinant protein bands should appear after IPTG induction (Figure 1A).
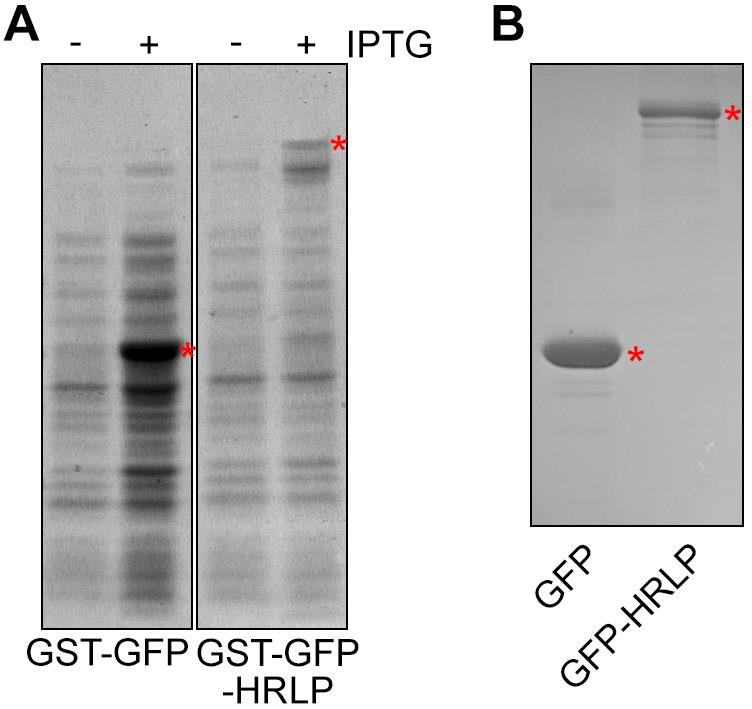
Figure 1. Protein staining with Coomassie blue. (A) GST-GFP and GST-GFP-HRLP expression after IPTG induction. Red asterisks indicate the protein bands of GST-GFP and GST-GFP-HRLP. (B) GFP and GFP-HRLP proteins after cleavage. The red asterisks indicate the protein bands of GFP and GFP-HRLP.
Recombinant protein purification
After confirming the solubility of proteins, harvest the induced cells containing GST-GFP and GST-GFP-HRLP by centrifugation at 2,400× g for 5 min at 4 °C.
Resuspend the cells in 10 mL of ice-cold lysis buffer, followed by sonication (output watt at 6, 30 s on/ 30 s off) until the cell suspension becomes clear.
Centrifuge the suspension at 10,000× g for 20 min at 4 °C.
Transfer the supernatant containing induced proteins into new 15 mL Falcon tubes.
Equilibrate the Glutathione Sepharose beads by washing the beads with 1 mL of lysis buffer.
Transfer 150 μL of equilibrated beads to each supernatant, followed by an incubation for 2 h at 4 °C with gentle rotation.
Centrifuge at 200× g for 1 min at 4 °C to pellet the beads.
Discard the supernatants and wash beads with 5 mL of ice-cold lysis buffer for 5 min.
Repeat the washing step twice.
To test the protein quality and size, boil 2 μL of washed beads in 10 μL of lysis buffer supplemented with 6× SDS loading dye at 100 °C for 10 min and run in an SDS-PAGE gel.
After running is completed, stain the gel with Coomassie blue buffer for 1 h at room temperature.
Destain the gel with destaining solution; the target protein bands should appear after IPTG induction. For the rest of the beads, keep at 4 °C until the cleavage step.
Cleavage of recombinant protein
Add 5 μL of PreScission Protease and 200 μL of cleavage buffer to the washed beads.
Incubate the beads on a nutating mixer at 4 °C for 4 h (Note 3).
Centrifuge the beads at 600× g for 1 min at 4 °C.
Transfer the green supernatants to new 1.5 mL tubes to store GFP and GFP-HRLP proteins (Figure 2). The proteins can be stored in cleavage buffer in small aliquots at -80 °C for several months (Note 4).
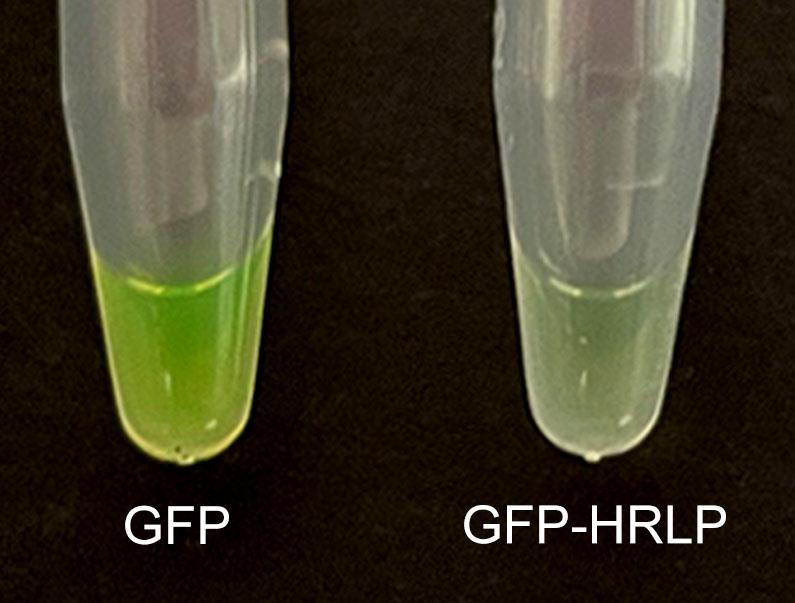
Figure 2. GFP (left) and GFP-HRLP (right) proteins after cleavageMeasure the protein concentration with Bio-Rad protein assay dye reagent concentrate (BSA protein could be used to generate the standard curve).
Run 2 μL of GFP and GFP-HRLP proteins on a protein gel followed by Coomassie blue staining to check the protein quality and size (Figure 1B) (Note 5).
In vitro phase separation
Dilute the generated GFP and GFP-HRLP proteins at the final concentration of 10 μM (Note 6) with the cleavage buffer on ice.
Add 2 μL of PEG-8000 (Note 7) at the final concentration of 10% to the 18 μL of diluted proteins in a PCR tube on ice. Mix the samples by pipetting up and down.
Transfer the 20 μL mixture to a Hole microscope slide immediately to avoid protein degradation or fluorescence decay.
Carefully cover the slide with a cover glass.
Observe the fluorescence with 40× microscope objective under an FV3000 Olympus confocal microscope. GFP protein exhibits uniform signals under the microscope, whereas GFP-HRLP forms droplets (Figure 3) (Notes 8–10).
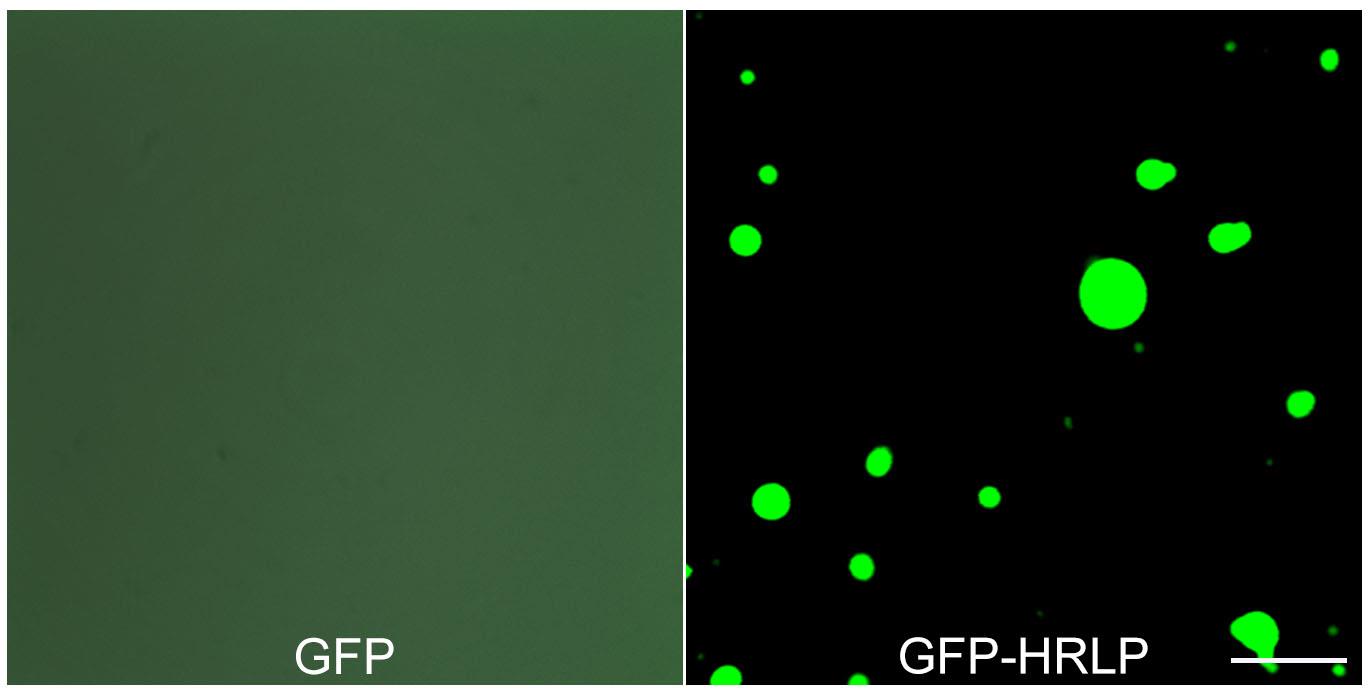
Figure 3. In vitro phase separation of GFP and GFP-HRLP proteins with the addition of PEG-8000. Scale bars, 10 μm.
Data analysis
The in vitro phase separation assay should be repeated through the above procedures using independently expressed and purified recombinant proteins. The negative control protein GFP should always be included. If a protein of interest but not the GFP control consistently form droplets in several independent experiments, this protein of interest undergoes LLPS in vitro. Other methods including timelapse imaging and FRAP are needed to further confirm the LLPS property of the protein of interest.
Notes
After IPTG induction, the color of the cell culture usually turns green, which is an indicator of successful induction of GFP fusion proteins.
The concentration of NaCl in the lysis buffer could be adjusted based on the specific properties of the protein of interest, as certain phase-separating proteins may require higher concentrations of NaCl to enhance their solubility. This adjustment is aimed at optimizing the conditions for maintaining the protein of interest in a soluble state.
The concentration of PreScission protease and the duration of incubation time could be adjusted on a case-by-case basis to avoid protein degradation.
The protein aliquots should be stored in small volumes, and repeated freeze-thaw cycles should be avoided to maintain the integrity and activity of proteins.
A size-exclusion chromatography assay is suggested to effectively eliminate nonspecific proteins and contaminants.
Optimal protein concentrations should be determined on a case-by-case basis. It is recommended to evaluate whether liquid droplet formation is concentration dependent.
The concentrations of PEG-8000 could be adjusted. Other crowding reagents, such as dextran and Ficoll, could also be used.
Some proteins may need longer time to form droplets.
The formation of protein droplets is affected by multiple factors such as pH, salt concentration, protein concentration, temperature, and PEG concentration. Modifying these parameters may promote droplet formation.
The presence of a GFP tag may influence the phase separation properties of certain proteins. In such cases, it is recommended to remove the GFP tag and conduct LLPS observations under a Differential Interference Contrast (DIC) microscopy. DIC microscopy allows for the visualization of phase-separated structures without the potential interference from the GFP tag, providing a clearer assessment of a protein's inherent phase separation behavior.
Acknowledgments
This work was supported by the National Research Foundation Competitive Research Programme (NRF-CRP22-2019-0001) and the intramural research support from Temasek Life Sciences Laboratory.
This protocol was derived from the original work of Zhang et al. (2022).
Competing interests
The authors declare no competing interests.
References
- Boeynaems, S., Alberti, S., Fawzi, N. L., Mittag, T., Polymenidou, M., Rousseau, F., Schymkowitz, J., Shorter, J., Wolozin, B., Van Den Bosch, L., et al. (2018). Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 28(6): 420–435.
- Dao, T. P., Kolaitis, R. M., Kim, H. J., O’Donovan, K., Martyniak, B., Colicino, E., Hehnly, H., Taylor, J. P. and Castañeda, C. A. (2018). Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 69(6): 965–978.e6.
- Dignon, G. L., Best, R. B. and Mittal, J. (2020). Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 71(1): 53–75.
- Li, J., Zhang, M., Ma, W., Yang, B., Lu, H., Zhou, F. and Zhang, L. (2022). Post-translational modifications in liquid-liquid phase separation: a comprehensive review. Mol. Biomed. 3(1): e1186/s43556-022-00075-2.
- Saha, S., Weber, C. A., Nousch, M., Adame-Arana, O., Hoege, C., Hein, M. Y., Osborne-Nishimura, E., Mahamid, J., Jahnel, M., Jawerth, L., et al. (2016). Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell 166(6): 1572–1584.e16.
- Zhang, Y., Fan, S., Hua, C., Teo, Z. W. N., Kiang, J. X., Shen, L. and Yu, H. (2022). Phase separation of HRLP regulates flowering time in Arabidopsis. Sci. Adv. 8(25): eabn5488.
Article Information
Publication history
Published: Sep 5, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, Y. and shen, L. (2023). An in vitro Assay to Probe the Formation of Biomolecular Condensates. Bio-protocol 13(17): e4813. DOI: 10.21769/BioProtoc.4813.
- Zhang, Y., Fan, S., Hua, C., Teo, Z. W. N., Kiang, J. X., Shen, L. and Yu, H. (2022). Phase separation of HRLP regulates flowering time in Arabidopsis. Sci. Adv. 8(25): eabn5488.
Category
Biochemistry > Protein > Fluorescence
Plant Science > Plant biochemistry > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







