- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescence in situ Localization of Pri-miRNAs in Isolated Arabidopsis thaliana Nuclei
Published: Vol 13, Iss 18, Sep 20, 2023 DOI: 10.21769/BioProtoc.4824 Views: 364
Reviewed by: Durai SellegounderEduardo ListikShreya MukhopadhyayAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
A Novel Method to Isolate RNase MRP Using RNA Streptavidin Aptamer Tags
Violette Charteau [...] Ger J. M. Pruijn
Feb 20, 2023 823 Views
An Optimized Protocol for Detecting Guard Cell–specific Gene Expression by in situ RT-PCR in Brassica rapa
Yingying Song [...] Xiaowu Wang
Sep 5, 2023 272 Views

Absolute Quantification of mRNA Isoforms in Adult Stem Cells Using Microfluidic Digital PCR
Shubhangi Das Barman [...] Antoine de Morree
Sep 5, 2023 260 Views
Abstract
Here, we present an approach combining fluorescence in situ hybridization (FISH) and immunolabeling for localization of pri-miRNAs in isolated nuclei of A. thaliana. The presented method utilizes specific DNA oligonucleotide probes, modified by addition of digoxigenin-labeled deoxynucleotides to its 3′ hydroxyl terminus by terminal deoxynucleotidyl transferase (TdT). The probes are then detected by immunolabeling of digoxigenin (DIG) using specific fluorescent-labeled antibodies to visualize hybridized probes. Recently, we have applied this method to localize pri-miRNA156a, pri-miRNA163, pri-miRNA393a, and pri-miRNA414 in the nuclei isolated from leaves of 4-week-old A. thaliana. The present approach can be easily implemented to analyze nuclear distribution of diverse RNA classes, including mRNAs and pri-miRNAs in isolated fixed cells or nuclei from plant.
Keywords: Fluorescent in situ hybridizationBackground
Microscopic imaging allows to observe and analyze the localization of diverse RNA molecules in their native cellular environment. It was found to be essential for understanding their turnover and function in different types of cells. Conventional fluorescence in situ hybridization (FISH) techniques have been designed for visualization of RNAs and relay on using fluorescently labeled DNA oligonucleotides (Hu et al., 2014). However, FISH methods are often challenging to detect RNAs expressed at low levels in the cells, such as pri-miRNAs, mainly due to low efficiency of resulting fluorescence signal. Here, we describe a method to detect pri-miRNA molecules in isolated nuclei of A. thaliana by combining FISH together with immunolocalization of digoxigenin (Figure 1). This method has been shown to be more sensitive and effective than conventional FISH (Bhat et al., 2020; Gonzalo et al., 2022; Stepien et al., 2022), and can successfully be applied to detect pri-miRNAs in plant and animal cells.
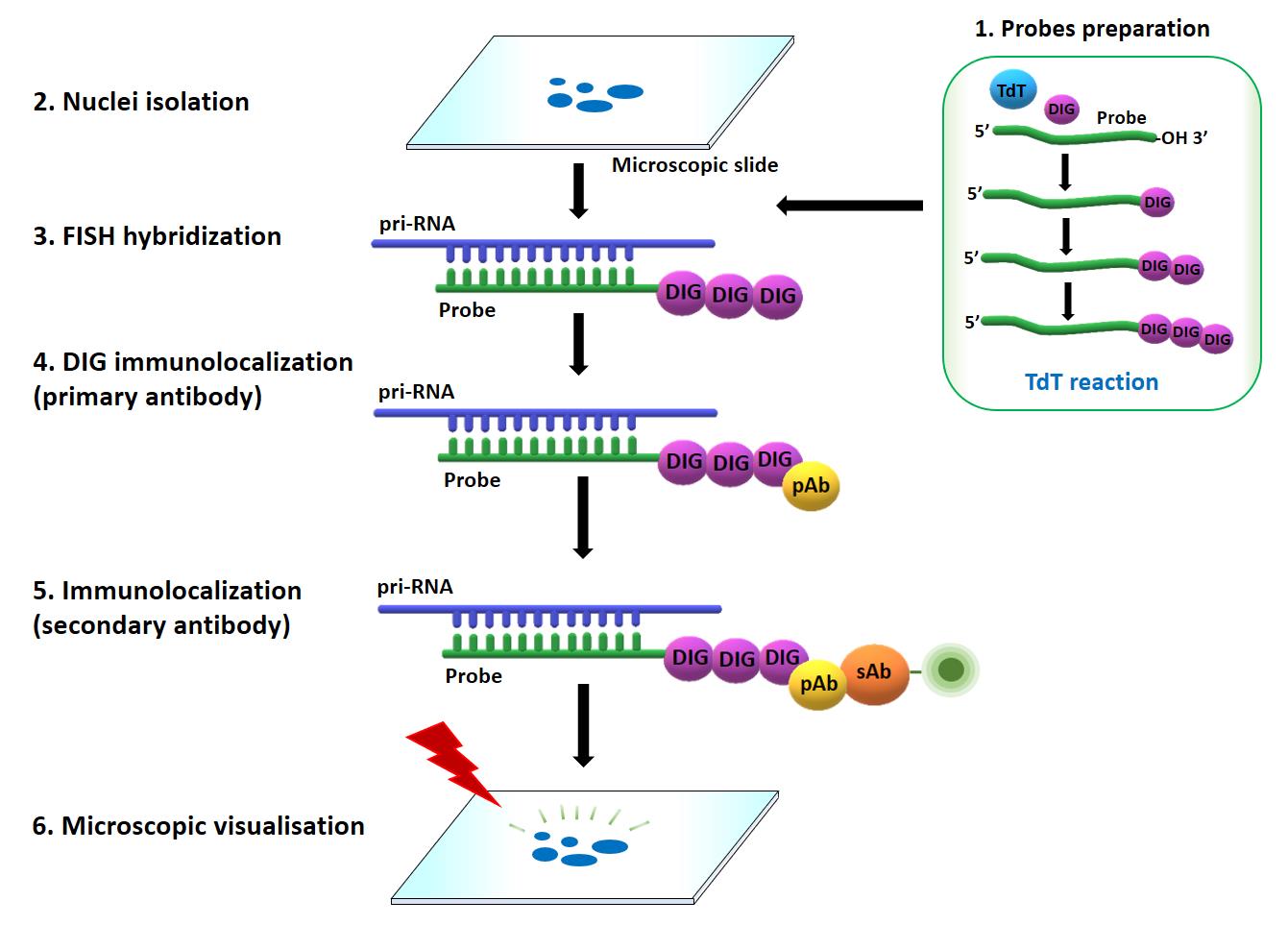
Figure 1. Overview of combined fluorescence in situ hybridization (FISH) and immunolabeling approach to detect pri-miRNAs in situ. Prior to the FISH, DNA oligonucleotides were labeled with digoxigenin at their 3′-ends by terminal transferase (TdT) (1), and the nuclei were isolated following the procedure described in (2). Pri-miRNAs were next localized by applying FISH (3) combined with the immunolocalization of digoxigenin attached to 3′-ends of the probes (4 and 5). The results were registered with confocal microscope (6). DIG: digoxigenin, TdT: terminal transferase, pAb: primary antibody, sAb: secondary antibody
Materials and reagents
Reagents
Roche Terminal Transferase 8000 U (Merck, catalog number: 3333566001)
ChromaTideTM Alexa FluorTM 488-5-dUTP (Thermo Fisher, catalog number: C11397)
Roche DIUTPS-RO (DIG-11-dUTP) (Digoxigenin-11-dUTP, alkali-stable) (Merck, catalog number: 11093088910)
Hoechst 33342, trihydrochloride, trihydrate, 100 mg (Thermo Fisher, catalog number: H1399)
ProLongTM Gold Antifade Mountant (Thermo Fisher, catalog number: P36930)
Paraformaldehyde, 16% w/v aq. soln., methanol free (Thermo Fisher, catalog number: 043368.9M)
Formamide (Merck, catalog number: FX0420)
Sodium chloride (Merck, catalog number: 1.16224)
Potassium chloride (Merck, catalog number: 7447-40-7)
Magnesium chloride (Merck, catalog number: 7786-30-3)
Sodium citrate (Merck, catalog number: 1613859)
Sodium phosphate, 0.5 M, pH 7.0 buffer (Thermo Fisher, catalog number: J63791.AK)
Saline sodium citrate (SSC) 20× concentrate (Merck, catalog number: S6639)
Ethylenediaminetetraacetic acid (EDTA) (Merck, catalog number: E9884-100G)
Bovine serum albumin (BSA) (Merck, catalog number: A9418-5G)
Ficoll® 400 (Merck, catalog number: F2637-5G)
Polyvinylpyrrolidone (Merck, catalog number: P0930-50G)
BSA, acetylated (Thermo Fisher, catalog number: AM2614)
Spermine (Merck, catalog number: S3256-1G)
Anti-digoxigenin antibody (Merck, 11333062910)
Goat anti-mouse secondary antibody, Alexa Fluor Plus 488 (Thermo Fisher, catalog number: A55058)
Goat anti-mouse secondary antibody, Alexa Fluor Plus 555 (Thermo Fisher, catalog number: A32727)
RNase A (Thermo Fisher, catalog number: EN0531)
Tris-HCl at pH 7.5 (Thermo Fisher, catalog number: 15567027)
Tween 20 (Merck, catalog number: P1379)
Sucrose (Merck, catalog number: S0389)
Phosphate buffered saline (PBS) (Merck, catalog number: P4417)
Triton X-100 (Merck, catalog number: 9036-19-5
Solutions
Hybridization buffer (see Recipes)
Denhardt’s solution 100× (50 mL) (see Recipes)
LB01 buffer (10 mL) (see Recipes)
Sorting buffer (10 mL) (see Recipes)
Equipment
Humid chamber (e.g., StainTray Slide Staining, Polysciences, catalog number: 25502-1)
Polylysine adhesion slides (e.g., Roth, catalog number: ET10.1)
Cover glasses for microscope slides (e.g., Merck, Brand cover glasses, catalog number: BR470045)
CellTrics 30 µm (Sysmex, catalog number: CTS220112)
Confocal microscope, Leica DMI 6000B, TCS TrinoTCS SP8
Plan Apochromat DIC H oil immersion lens (63×/1.4)
Software
ImageJ, Fiji (https://imagej.net/software/fiji/)
Procedure
Nuclei isolation
Note: Nuclei isolation was performed according to the method described in Pontvianne et al. (2007 and 2012).
Collect 2–3 leaves from 4-week-old A. thaliana plants and fix them for 20 min at room temperature in 4% formaldehyde solution in Tris buffer (10 mM Tris-HCl at pH 7.5, 10 mM EDTA, 100 mM NaCl). Leaves should be completely immersed.
Notes:
For 4% fixative preparation, dilute 10 mL of 16% methanol-free formaldehyde in 30 mL of Tris buffer.
Nuclei can also be isolated from seedlings, following the same protocol.
WARNING: Formaldehyde is a volatile and toxic carcinogen and should only be used in a fume hood. Gloves, safety glasses, and protective clothing should also be worn when working with it.
Wash the leaves twice for 10 min in Tris buffer.
Chop the leaves with a razor blade in 0.5 mL of LB01 buffer to obtain a soup in a Petri dish.
Filter the resulting lysate through a 30 μm cell strainer.
Add 12 μL of sorting buffer per 3 μL of filtered nuclei suspension obtained in step 3.
Spread the mixture on adhesion slides covered with Polylysine using a pipette and leave to dry at room temperature (can be kept overnight).
Probe preparation
Dissolve DNA oligonucleotides with double-distilled H2O to a final concentration of 100 μM.
Prepare terminal transferase (TdT) working solution: 5 mM CoCl2, 1× TdT Reaction buffer, 0.1 mM DIG-11-dUTP, 0.1 mM dATP, and 400 U of TdT per reaction in PCR tube.
Notes:
Optionally, you can supply a reaction buffer additionally with Alexa Fluor 5-dUTP (final concentration: 0.2 mM).
Instead of DIG-11-dUTP, Biotin-11-dUTP could be used.
Combine 1 μL of DNA oligonucleotide with 9 μL of TdT working solution and incubate it for 40 min at 37 °C in a PCR tube.
Notes:
Incubation is performed without shaking. After incubation, the reaction mixture could be stored at -20 °C. In our studies, probes were working after three months of storage.
For our studies, the probes were provided by Metabion company (Planegg, Germany).
Fluorescence in situ hybridization (FISH)
Prepare permeabilization buffer: 0.1% Triton X-100 in 1× PBS.
Incubate the sample in permeabilization buffer for 10 min at room temperature. The buffer should cover the whole sample area.
Wash the sample twice with 1× PBS.
Prepare hybridization buffer (Niedojadlo et al., 2012).
For FISH, dilute 1 μL of the probe mixture (section B) in 100 μL of hybridization buffer (final probe concentration: 100 pM/μL).
Note: The final probe concentration in hybridization buffer may vary for different RNAs and should be adjusted experimentally.
Add 100 μL of hybridization buffer to the sample on the slide and cover with a slide cover.
Place the slide in a humid hybridization chamber in the 28 °C incubator and leave overnight. The slides should be prevented from drying during hybridization. To obtain this, a small volume of PBS or water should be placed inside the chamber to keep a stable humidity.
Note: Incubation is performed with a gentle shaking (0.6 rpm)
After incubation, wash the slide using the following order: 4× SSC for 5 min, 2× SSC for 5 min, 1× SSC for 5 min, and 1× PBS for 10 min.
Immunolabeling
Dilute primary anti-digoxigenin antibody in 1× PBS containing 0.05% acetylated BSA diluted 1:100.
Cover the sample with 100 μL of antibody diluent (containing anti-digoxigenin antibody from step 1) and place the slide cover. Incubate the sample in a humid chamber overnight at 10 °C.
Remove primary antibody from the sample and wash with 1× PBS for 5 min.
Dilute the secondary antibody labeled with fluorescence dye in 1× PBS containing 0.05% acetylated BSA (diluted 1:1,000).
Place 100 μL of secondary antibody solution on the sample, cover it with a slide cover glass, and incubate in a humid chamber for 3 h at 37 °C.
Remove the secondary antibody from the sample and wash it two times with 1× PBS for 5 min.
Note: The time and temperature of incubation, as well as antibody concentration, should be defined experimentally for different antibodies used.
Nuclei labeling and sample mounting
Dilute Hoechst 33342 at 1:1,000 in 1× PBS and add 100–200 μL to the sample. Incubate for 10 min at room temperature.
Note: For nuclei labeling, DAPI (4′,6-diamidino-2-phenylindole) diluted 1:1,000 in 1× PBS buffer can also be used.
Remove the Hoechst 33342 solution from the slide and wash it once with 1× PBS for 5 min.
Cover the sample with 20 μL of ProLong Gold Antifade reagent, carefully place the coverslip, and press down firmly with the finger to remove air bubbles and uniformly distribute the antifade reagent over the samples.
Leave the sample to dry at room temperature, protecting it from the light.
Data analysis
In our studies, the results were registered with the Leica SP8 confocal microscope using a diode 405 laser, an argon/ion laser with a wavelength of 488 nm, and a diode laser DPSS 561 that emitted light with a wavelength of 561 nm with an optimized pinhole, long exposure time (200 kHz) and 63× (numerical aperture, 1.4). Images were collected sequentially in blue (Hoechst 33342), green (Alexa 488 fluorescence), and red (Alexa 555) channels. The low laser power (0.4%–5% of maximum power) and a single-channel collection were set to minimize bleed-through between fluorescence channels. For bleed-through analysis, Leica SP8 software was used.
The presented approach was previously applied to localize pri-miRNA156a and pri-miRNA163 in the nuclei isolated from leaves of 4-week-old A. thaliana (Figure 2).
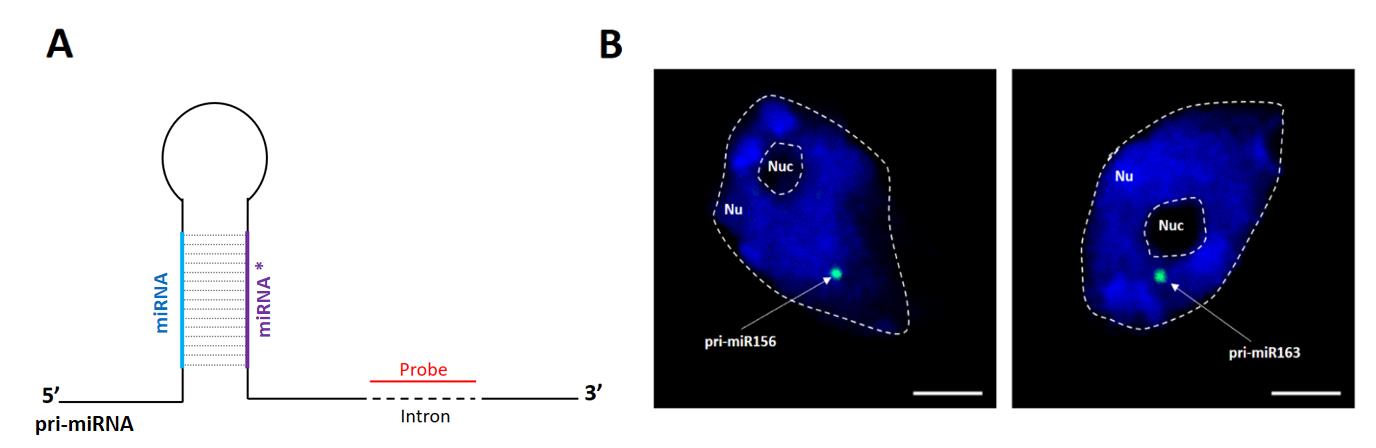
Figure 2. Detection of pri-miR156a and pri-miR163 using digoxigenin-labeled probes and antibodies targeting digoxigenin. The probes hybridizing to intron sequence located downstream of stem-loop structure were applied. (A) Schematic illustration of stem-loop structure of pri-miRNA; probe is depicted in red. (B) Detection of pri-miRNA156a (left image, green) and pri-miRNA163 (right image, green). DNA was stained with Hoechst (blue). Scale bar = 2.5 μm.
To confirm that the observed nuclear spots were due to hybridization of probes to pri-miRNA, we treated the isolated nuclei with RNase A (100 μg/mL in PBS for 30 min) and then applied the probe recognizing the intronic sequence of pri-miR156a (Figure 3B). We have not observed hybridization signals in nuclei treated with RNase A. Similarly, we have not observed fluorescence signal if a sense probe matching the pri-miR156a intron was applied (Figure 3C). Additionally, we have calculated a fluorescence intensity by Fiji software to distinguish the positive fluorescence signal of hybridization from the background.
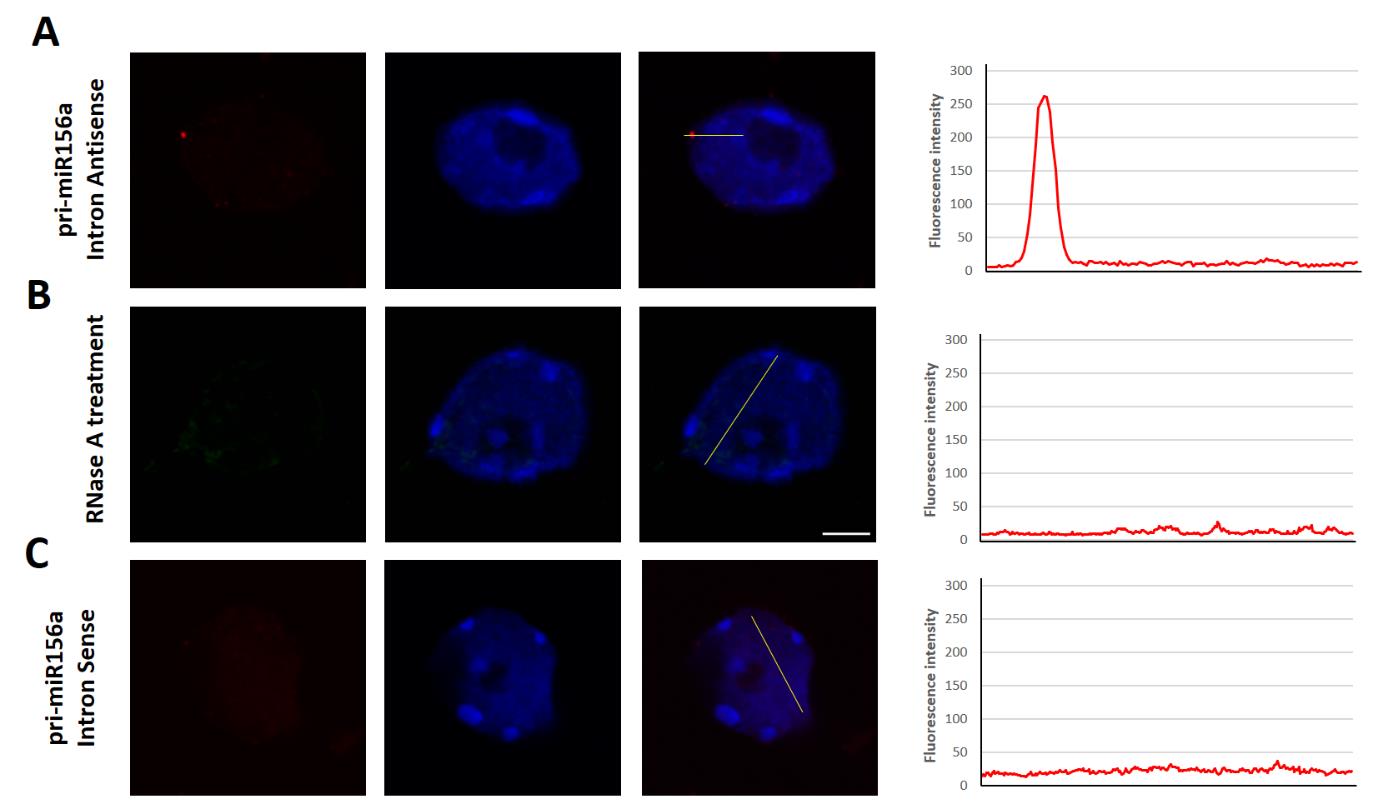
Figure 3. Detection (fluorescence in situ hybridization, FISH) of pri-miR156a (red) using the intron-recognizing probe (Intron antisense probe), in the nuclei treated with RNase A and using the probe with the sequence identical with a fragment of intron (Intron sense probe). The fluorescence intensity is plotted along the yellow line shown on each merged image. Scale bar = 2.5 μm.
Recipes
Hybridization buffer
Reagent Final concentration Quantity Formamide 30% (v/v) 600 μL 20× SSC 4× 400 μL 100× Denhardt’s solution 5× Denhardt’s buffer 100 μL Sodium phosphate buffer (0.5 M) 50 mM 200 μL EDTA (0.5 M) 1 mM 4 μL H2O n/a 696 μL Total n/a 2 mL Denhardt’s solution 100× (50 mL)
Reagent Final concentration Quantity Bovine serum albumin (BSA) 2% (w/v) 1 g Ficoll 400 2% (w/v) 1 g Polyvinylpyrrolidone 2% (w/v) 1 g H2O n/a 50 mL Total n/a 50 mL LB01 buffer (10 mL)
Reagent Final concentration Quantity Tris-HCl (1M) 15 mM 150 μL EDTA 2 mM 20 μL Spermine 0.5 mM 5 μL Potassium chloride (KCl) (1M) 80 mM 800 μL Sodium chloride (NaCl) (1M) 20 mM 200 μL Triton (10%) 0.1% 1 mL Sorting buffer (10 mL)
Reagent Final concentration Quantity Tris-HCl (1 M) 100 mM 1 mL Potassium chloride (KCl) (1 M) 50 mM 500 μL Magnesium chloride (MgCl2) (1 M) 2 mM 20 μL Tween 20 (10%) 0.05 mM 50 μL Sucrose 5% 0.5 g Bovine serum albumin (BSA) 3% 0.3 g
Acknowledgments
This work was supported by grants from the Polish National Science Centre (UMO-2019/32/T/NZ1/00508, UMO-2016/23/N/NZ1/00010, UMO-2013/10/A/NZ1/00557) and the Initiative of Excellence–Research University (05/IDUB/2019/94) at Adam Mickiewicz University, Poznan, Poland, to A.J.
Competing interests
There are no conflicts of interest or competing interests.
References
- Bhat, S. S., Bielewicz, D., Gulanicz, T., Bodi, Z., Yu, X., Anderson, S. J., Szewc, L., Bajczyk, M., Dolata, J., Grzelak, N., et al. (2020). mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 117(35): 21785–21795.
- Gonzalo, L., Tossolini, I., Gulanicz, T., Cambiagno, D. A., Kasprowicz-Maluski, A., Smolinski, D. J., Mammarella, M. F., Ariel, F. D., Marquardt, S., Szweykowska-Kulinska, Z., et al. (2022). R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants. Nat. Plants 8(4): 402–418.
- Hu, L., Ru, K., Zhang, L., Huang, Y., Zhu, X., Liu, H., Zetterberg, A., Cheng, T. and Miao, W. (2014). Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. Biomarker Res. 2(1): e1186/2050-7771-2-3.
- Niedojadlo, K., Piecinski, S., Smolinski, D. J., Bednarska-Kozakiewicz, E. (2012) Transcriptional activity of Hyacinthus orientalis L. female gametophyte cells before and after fertilization. Planta 236: 153–169.
- Pontvianne, F., Matía, I., Douet, J., Tourmente, S., Medina, F. J., Echeverria, M. and Sáez-Vásquez, J. (2007). Characterization of AtNUC-L1 Reveals a Central Role of Nucleolin in Nucleolus Organization and Silencing of AtNUC-L2 Gene in Arabidopsis. Mol. Biol. Cell 18(2): 369–379.
- Pontvianne, F., Blevins, T., Chandrasekhara, C., Feng, W., Stroud, H., Jacobsen, S. E., Michaels, S. D. and Pikaard, C. S. (2012). Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26(9): 945–957.
- Stepien, A., Dolata, J., Gulanicz, T., Bielewicz, D., Bajczyk, M., Smolinski, D. J., Szweykowska-Kulinska, Z. and Jarmolowski, A. (2022). Chromatin-associated microprocessor assembly is regulated by the U1 snRNP auxiliary protein PRP40. Plant Cell 34(12): 4920–4935.
Article Information
Publication history
Published: Sep 20, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Gulanicz, T., Zienkiewicz, A., Zienkiewicz, K., Kasprowicz-Maluski, A., Szweykowska-Kulinska, Z. and Jarmolowski, A. (2023). Fluorescence in situ Localization of Pri-miRNAs in Isolated Arabidopsis thaliana Nuclei. Bio-protocol 13(18): e4824. DOI: 10.21769/BioProtoc.4824.
Category
Molecular Biology > RNA > RNA detection
Plant Science > Plant molecular biology > RNA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







