- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Human Dendritic Cell Subset Isolation by Magnetic Bead Sorting: A Protocol to Efficiently Obtain Pure Populations
Published: Vol 13, Iss 20, Oct 20, 2023 DOI: 10.21769/BioProtoc.4851 Views: 352
Reviewed by: Chiara AmbrogioWendy Leanne Hempstock

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Isolation of Stem Cells, Endothelial Cells and Pericytes from Human Infantile Hemangioma
Lan Huang and Joyce Bischoff
Jan 20, 2020 3405 Views
Isolation of CD31+ Bone Marrow Endothelial Cells (BMECs) from Mice
Alhaji Osman Smith [...] Lingyu Zeng
Nov 20, 2021 2301 Views
A Rapid Protocol for Direct Isolation of Osteoclast Lineage Cells from Mouse Bone Marrow
Lei Dang [...] Jin Liu
Mar 5, 2022 2608 Views
Abstract
Dendritic cells have been investigated for cell-based immunotherapy for various applications. The low abundance of dendritic cells in blood hampers their clinical application, resulting in the use of monocyte-derived dendritic cells as an alternative cell type. Limited knowledge is available regarding blood-circulating human dendritic cells, which can be divided into three subsets: type 2 conventional dendritic cells, type 1 conventional dendritic cells, and plasmacytoid dendritic cells. These subsets exhibit unique and desirable features for dendritic cell-based therapies. To enable efficient and reliable human research on dendritic cell subsets, we developed an efficient isolation protocol for the three human dendritic cell subsets, resulting in pure populations. The sequential steps include peripheral blood mononuclear cell isolation, magnetic-microbead lineage depletion (CD14, CD56, CD3, and CD19), and individual magnetic-microbead isolation of the three human dendritic cell subsets.
Graphical overview
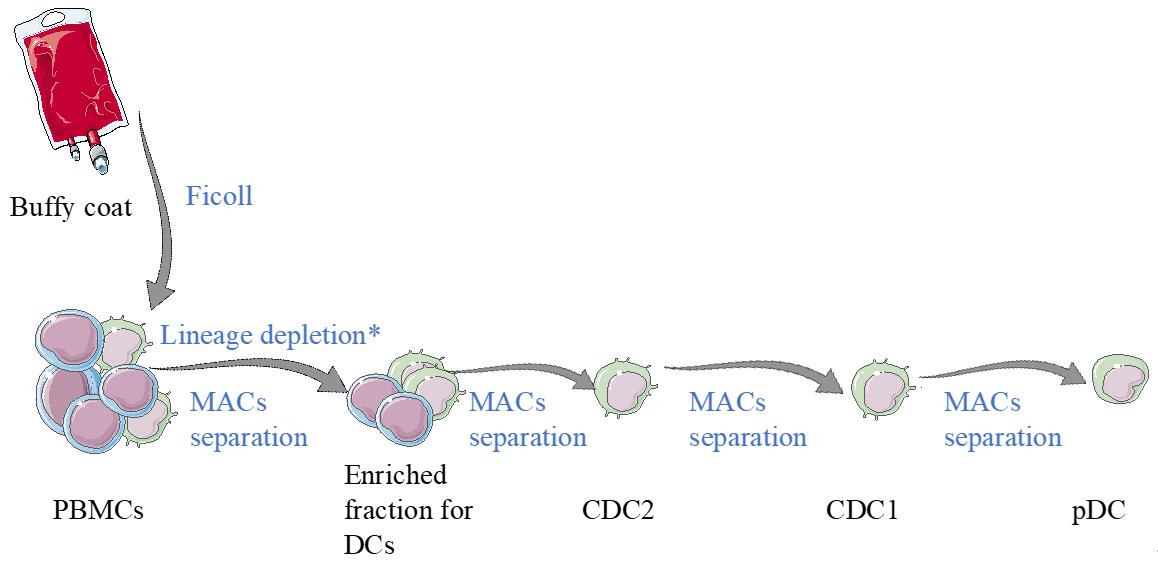
Scheme of the dendritic cell (DC) isolation protocol. Starting material for this process is human blood (buffy coat or aphaeresis). From that, peripheral blood mononuclear cells (PBMCs) are isolated by using ficoll gradient centrifugation. Then, an enrichment for DCs is performed using semi-automated equipment. From the enriched fraction, DC subsets are obtained by magnetic cell sorting.
Background
During the past decades, dendritic cells (DCs) have been investigated for their ability to initiate antigen-specific immune responses in vivo. DC-based immunotherapies have been developed or are under investigation for the treatment of various malignancies such as cancer (Gonzalez et al., 2018), autoimmune disorders such as rheumatoid arthritis, or multiple sclerosis (Collin and Bigley, 2018). Most of the current DC knowledge is from the human DC model, monocyte-derived DCs (moDCs), which have a very high purity and can be generated in significant numbers from peripheral blood mononuclear cells (PBMCs).
Recently, attention shifted towards the utilization of the three human DC subsets circulating in blood, due to their unique and distinct functions compared with moDCs (Wang et al., 2020). Blood-circulating human DCs can be broadly divided into plasmacytoid DCs (pDCs) and conventional DCs (cDCs). cDCs are highly specialized in antigen uptake, processing, and (cross-)presenting of antigens to naïve T cells, which is a crucial step for initiating immune responses. cDCs can be subdivided into type 1 (cDC1s) and type 2 (cDC2s). In humans, cDC1s express CD141 (BDCA-3), cDC2s express CD1c (BDCA-1), and pDCs express CD123 (BDCA-2). Each DC subset presents a unique function; for example, cDC1s can potently take up apoptotic cells, cross-present externally derived antigens, and activate cytotoxic lymphocytes (Schreibelt et al., 2012). These features place cDC1s in the center of interest for mounting anti-tumor immune responses. Thus, it is necessary to investigate the phenotypical and functional differences between human DC subsets in depth, with the goal of improving DC-based therapies.
Unfortunately, investigating the human DC subsets is a rather tedious task, mainly due to their scarcity in PBMCs, ranging from < 0.2% of PBMCs in the case of cDC2s and pDCs to < 0.08% of PBMCs in the case of cDC1s (van Beek et al., 2020). In addition, currently available human DC isolation protocols result in varying levels of cell impurity or cross-contamination with other human DC subsets. Contaminating cells present in DC cultures can greatly influence the phenotypical and functional outcome of DC research (van Beek et al., 2020), hampering the elucidation of the role of individual human DC subsets.
We have developed a protocol to isolate highly pure populations of human DC subsets starting from PBMCs. Using the protocol described here, DC subsets can be isolated with higher purity and yield compared with currently available options such as cell sorting or magnetic-microbead kits without the lineage depletion step. The protocol consists of multiple steps including depletion of monocytes (CD14+), B cells (CD19+), T cells (CD3+), and NK cells (CD56+) from PBMCs with magnetic microbeads, followed by DC isolation using magnetic microbeads specific for each DC subset: cDC2s (CD1c+), cDC1s (CD141+), and pDCs (CD304+). We present the protocol with the use of semi-automated equipment (MultiMACS Cell24 Separator Plus, Miltenyi Biotec) that significantly reduces the time needed for the DC isolation process.
Materials and reagents
Beads and items for cell separation
Anti-CD3 microbeads (Miltenyi Biotec, catalog number: 130-050-101)
Anti-CD14 microbeads (Miltenyi Biotec, catalog number: 130-050-201)
Anti-CD19 microbeads (Miltenyi Biotec, catalog number: 130-097-055)
FcR blocking reagent (Miltenyi Biotec, catalog number: 130-059-901)
Anti-CD56 microbeads (Miltenyi Biotec, catalog number: 130-050-401)
Anti-CD1c biotin (Miltenyi Biotec, catalog number: 130-119-475)
Anti-biotin microbeads (Miltenyi Biotec, catalog number: 130-090-485)
Anti-CD141 microbeads (Miltenyi Biotec, catalog number: 130-090-532)
LD columns (Miltenyi Biotec, catalog number: 130-042-901)
MS columns (Miltenyi Biotec, catalog number: 130-042-201)
LS columns (Miltenyi Biotec, catalog number: 130-042-401)
Multi-24 column blocks (8×) (Miltenyi Biotec, catalog number: 130-095-691)
Single-well deep-well plates (Miltenyi Biotec, catalog number: 130-114-966)
Buffers and media
Human albumin (Sigma-Aldrich, catalog number: H0900000)
PBS (Life Technologies, catalog number: 14190-094)
Human serum (Sigma-Aldrich, catalog number: H4522-100ML)
X-VIVO15 hematopoietic cell culture medium (Life Technologies, catalog number: BE02-060Q)
UltraPureTM 0.5 M EDTA, pH 8.0 (Thermo Fisher, catalog number: 15575020)
Ammonium chloride (Sigma-Aldrich, catalog number: 213330)
Potassium bicarbonate (Sigma-Aldrich, catalog number: 237205)
Disodium EDTA (Sigma-Aldrich, catalog number: E4884)
Bovine serum albumin (Thermo Fisher, catalog number: 23209)
Sodium azide (Merck, catalog number: RTC0000068- 1L)
Trypan Blue (Sigma-Aldrich, catalog number: T6146-25G)
Diluting buffer (see Recipes)
ACK lysis buffer (see Recipes)
Washing buffer (see Recipes)
PBA (see Recipes)
Antibodies
Anti-CD20 FITC (BD Biosciences, catalog number: 345792)
Anti-CD14 PerCP (BioLegend, catalog number: 325632)
Anti-CD141 APC (Miltenyi Biotec, catalog number: 130-090-907)
Anti-CD1c PE (Miltenyi Biotec, catalog number: 130-113-864)
Anti-CD123 APC (BD Biosciences, catalog number: 560087)
Anti-BDCA2 PE (Miltenyi Biotec, catalog number: 130-097-929)
Materials
Ficoll lymphoprep (VWR, catalog number: CLUT1114547)
Tuerk’s solution for leukocyte counting (Merck, catalog number: 1092770100)
Tubes (Stem Cell Technologies, catalog number: 100-0088)
50 mL tubes (Corning, catalog number: 430828)
96-well V-bottomed plate (Thermo Fisher, catalog number: 277143)
Microbeads, antibodies, X-VIVO15 hematopoietic cell culture medium, washing buffer, and PBA were stored at 4 °C. Diluting buffer, PBS, and ACK lysis buffer were stored at RT.
Note: Products and equipment from other vendors (when available) can also be used.
Equipment
MultiMACS Cell24 Separator Plus (Miltenyi Biotec, catalog number: 130-098-637)
QuadroMACS Separator (Miltenyi Biotec, catalog number: 130-090-976)
OctoMACS Separator (Miltenyi Biotec, catalog number: 130-042-109)
FACSVerse (Becton Dickinson)
Note: This FACS has three lasers and eight colors (four in the blue laser, two in the red laser, and two in the violet laser).
Centrifuge (Hettich centrifuge, model: rotanta 460R)
Cell counting chamber (Thermo Fisher, catalog number: C10228)
Shaker (Thermo Fisher, SHKE4000-7: MaxQ)
Software
FACS verse software BD (BD FACSVerseTM Systems)
FlowJo (FlowJoTM v10.8)
Procedure
From a buffy coat/aphaeresis:
Peripheral blood mononuclear cell (PBMC) isolation
Isolate PBMCs following a ficoll-based protocol (Corkum et al., 2015).
Note: From the PBMCs isolation, a cell suspension (with a mixture of different immune cells) is obtained.
After washing the cells three times with 1 mL of washing buffer (PBS, 0.1% BSA, EDTA), count the total amount of isolated PBMCs. For accurate counting, use Tuerk’s dye.
Notes:
In this protocol, washing the cells refers to centrifuging at 252× g for 5 min at 4 °C.
Counting is performed by using a cell counting chamber and a microscope, as shown in Figure 1.

Figure 1. Schematic protocol for how to count cellsResuspend PBMCs in 1 mL of 1× ACK lysis buffer per 100 × 106 cells.
Note: Resuspend by pipetting up and down.
Incubate for 5 min at room temperature (RT).
Add washing buffer to the ACK-cell suspension, filling up to 50 mL.
Note: Resuspend by pipetting up and down.
Centrifuge PBMCs at 252× g for 5 min at 4 °C. After centrifugation, the cells are in a pellet at the bottom.
Discard supernatant (by pouring it).
Proceed with the cell pellet to Section B.
Note: The cell pellet is resuspended by adding the lineage depletion mixture (Section B), followed by up-and-down pipetting.
Lineage depletion
Prepare the lineage depletion master mixture (Caution: This mixture is prepared using the PBMC count numbers from step A2):
Anti-CD3 microbeads at 1 μL per 1 × 106 PBMCs.
Anti-CD14 microbeads at 1 μL per 1 × 106 PBMCs.
Anti-CD19 microbeads at 1 μL per 1 × 106 PBMCs.
Anti-CD56 microbeads at 0.5 μL per 1 × 106 PBMCs.
Anti-FCR blocking at 1 μL per 1 × 106 PBMCs.
Add the depletion master mixture to the PBMC pellet and resuspend by pipetting up and down.
Incubate for 30 min at 4 °C while shaking on a shaker or manually every 5 min.
Note: Select the slowest speed of the shaker.
Add 5 mL of washing buffer to depletion mixture PBMC suspension (step B2).
Centrifuge PBMC suspension at 252× g for 5 min at 4 °C.
Discard the supernatant (by pouring it).
Resuspend the PBMCs at a concentration of 100 × 106 cells per milliliter in washing buffer.
Note: Resuspend by pipetting up and down.
Prepare the MultiMACS Cell24 Separator Plus without the elution station according to the manufacturer instructions. Initiate the preprogrammed depletion protocol.
Prewet the Multi-24 column block with 1 mL of washing buffer. Only prewet the number of columns required. Use one column per 100 × 106 PBMCs.
Place a single-well deep-well plate to recover the flowthrough containing the unlabeled cells (see Figure 2, which illustrates how the cells are collected in the system).
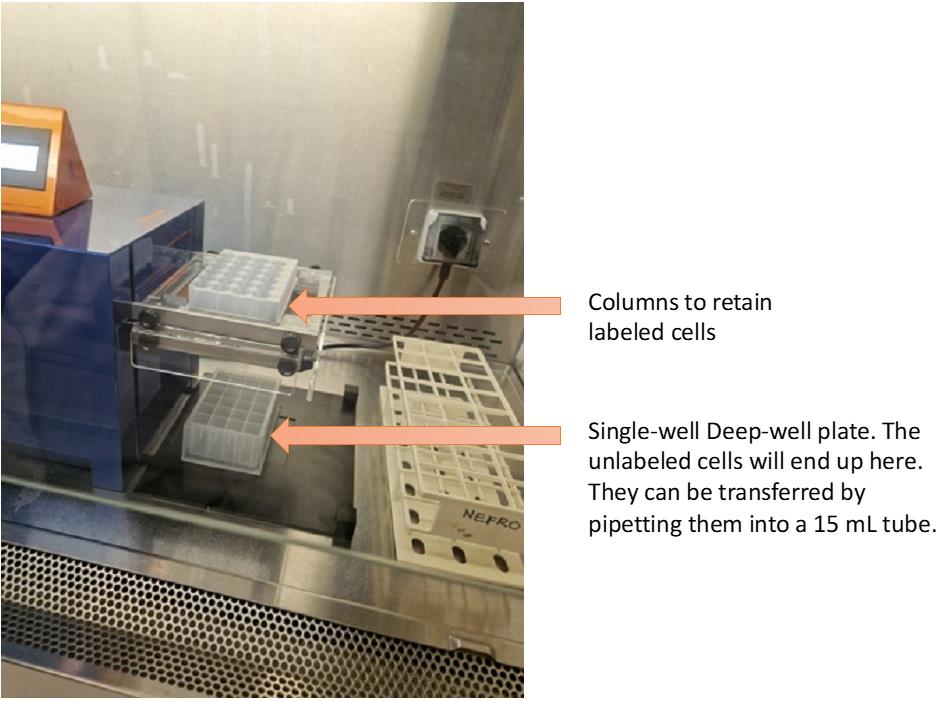
Figure 2. MultiMACS with the column block and the collectorFollow the indicated steps from the depletion protocol from the MultiMACS. In short, apply the PBMC suspension as 1 mL per column (Caution: The number of cells per column should be 100 × 106 PBMCs maximum). After the sample has run through the column, wash three times with 1 mL of washing buffer to ensure complete elution of target cell population.
Note: Washing the columns means adding the indicated buffer on top of the column and waiting until the fluid flows completely through.
Transfer the negative fraction (flowthrough) to a clean 50 mL tube. Centrifuge at 252× g for 5 min at 4 °C.
Note: The enriched DC fraction is the negative fraction.
Count the number of cells present in the negative fraction (with Tuerk’s dye and counting chamber).
Centrifuge at 252× g for 5 min at 4 °C.
Discard the supernatant (by pouring it).
Resuspend the pelleted cells at a concentration of 100 × 106 cells per milliliter with washing buffer.
Proceed to the final depletion step using LD columns.
Note: This step will ensure the highest purity of the negative fraction containing all DC subsets and a minute amount of other immune cells.
Prewet LD columns with 1 mL of washing buffer. Use 1 LD column per 100 × 106 cell suspension (step B13).
Run 1 mL of cell suspension per LD column and wash three times with 1 mL of washing buffer.
Collect negative fraction containing the target cell population (in a 15 mL tube, as shown in Figure 2).
Count the cells (using the counting chamber and Tuerk’s solution).
Centrifuge at 252× g for 5 min at 4 °C.
Discard the supernatant (by pouring it) and proceed with the cell pellet. The resuspension of the cell pellet will be done with the antibody mixture in Section C.
Continue with the isolation of the DC subsets.
cDC2 isolation
Add 1 μL of anti-CD1c biotin per 1 × 106 cells.
Note: Scale according to the starting cell number (step B19).
Incubate for 10 min at 4 °C while shaking.
Note: Select the lowest speed of the shaker.
Fill the 50 mL tube with washing buffer up to 50 mL.
Centrifuge at 252× g for 5 min at 4 °C.
Discard the supernatant.
Add 2 μL of anti-biotin microbeads per 1 × 106 cells.
Incubate for 15 min at 4 °C while shaking.
Note: Select the lowest speed of the shaker.
Add washing buffer up to 50 mL.
Centrifuge at 252× g for 5 min at 4 °C.
During centrifugation, prewet LS column with 1 mL of washing buffer.
Resuspend the cells in 1 mL of washing buffer.
Run the cells through the column that is placed in the magnet and wash three times with 1 mL of washing buffer.
Collect the flowthrough in a 50 mL tube (as shown in Figure 3).
Note: For the flushing step, the column should be removed from the magnet and placed in a 15 mL tube.
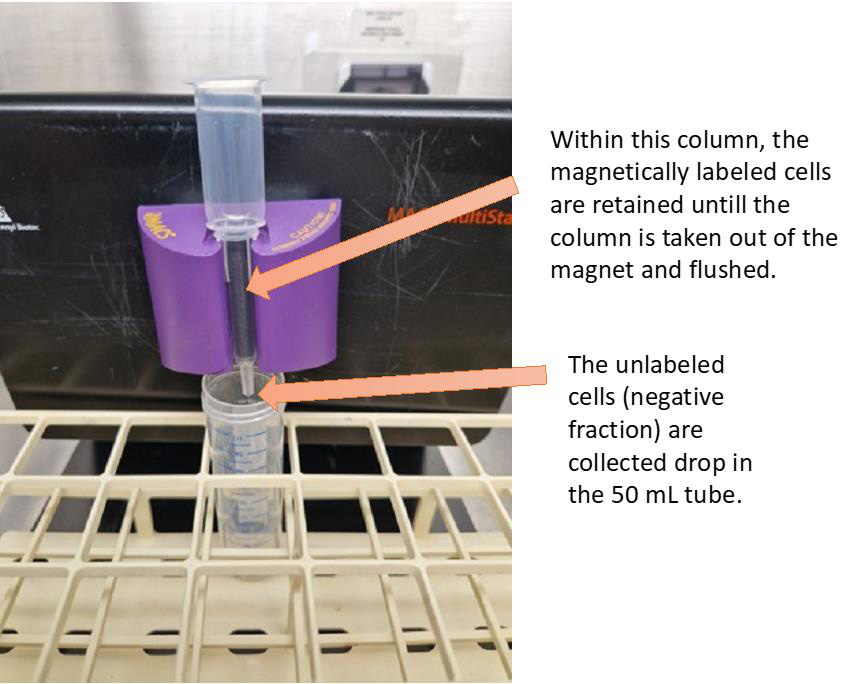
Figure 3. Setup of the LS column within the magnet; unlabeled cells go through, and labeled cells are retained by the magnetic field. Flush the LS column with 2 mL of washing buffer. Caution: This fraction contains the cDC2 population.To further increase the purity of the cDC2 population, prewet one MS column with 1 mL of washing buffer.
Run the 2 mL of cDC2 cell suspension through the MS column. Wash three times with 0.5 mL of washing buffer.
Flush cDC2s out of the MS column with 2 mL of washing buffer.
Count the cDC2s (using trypan blue solution and a counting chamber, as illustrated in Figure 1).
Resuspend the cells (cDC2s) at 1 × 106 cells per milliliter with X-VIVO15 + 2% human serum and keep them at 4 °C.
cDC1 isolation
Centrifuge the flowthrough (unlabeled negative fraction) of the cDC2 isolation step (step C12) at 252× g for 5 min at 4 °C.
Discard the supernatant (by pouring it).
Count the cells (using the counting chamber and Tuerk’s solution).
Add 2 μL of anti-CD141 microbeads per 1 × 106 cells.
Incubate for 15 min at 4 °C while shaking.
Add washing buffer up to 50 mL.
Centrifuge at 252× g for 5 min at 4 °C.
During centrifugation, prewet one LS column with 1 mL of washing buffer.
Resuspend cell fraction in 1 mL.
Load the cells onto the LS column and wash the column three times with 1 mL of washing buffer.
Collect the flowthrough in a Falcon tube (50 or 15 mL).
Flush the LS column with 2 mL of washing buffer. Caution: This fraction contains the cDC1 population.
Note: For the flushing step, the column should be removed from the magnet and placed in a 15 mL tube.
To further increase the purity of the cDC1 population, prewet one MS column with 1 mL of washing buffer.
Run the 2 mL of cDC1s through the MS column that is placed in the magnet. Wash three times with 0.5 mL of washing buffer.
Flush cDC1s out of the MS column with 2 mL of washing buffer.
Count the cDC1 fraction (using trypan blue and the counting chamber).
Resuspend the cells at 1 × 106 cells per milliliter in X-VIVO15 + 2% human serum and keep them at 4 °C.
pDC isolation
Centrifuge the flowthrough (unlabeled negative fraction) from the cDC1 isolation (step D9) at 252× g for 5 min at 4 °C.
Discard the supernatant (by pouring it).
Count the cells (using the counting chamber and Tuerk’s solution).
Add 1 μL of anti-CD304 microbeads per 1 × 106 cells.
Incubate for 15 min at 4 °C while shaking.
Add washing buffer up to 50 mL.
Centrifuge at 252× g for 5 min at 4 °C.
During centrifugation, prewet one LS column with 1 mL of washing buffer.
Resuspend the cells in 1 mL of washing buffer (PBS, 0.1% BSA, EDTA).
Pass the cells through the column (add the cell suspension on top of the column that is placed in the magnet).
Discard the flowthrough (as of now, it will no longer be needed).
Run the cells through the column that is placed in the magnet and add 1 mL of washing buffer. Flush the pDC out of the LS column by washing three times with 1 mL of washing buffer and count the cells. Caution: This fraction contains the pDC population.
Note: For the flushing step, the column should be removed from the magnet and placed in a 15 mL tube.
Resuspend the cells at 1 × 106 cells per milliliter in X-VIVO15 + 2% human serum and keep them at 4 °C.
Note: Cells can be cultured for up to 48 h with high percentage of viable cells.
Purity check
Note: Flow cytometry analysis can be performed the day after the isolation of the three DC subsets. Until the analysis is performed, cells should be stored at 4 °C.
Fill the wells of a 96-well V-bottomed plate with at least 10,000 cells/well (also considering the unstained control and isotype).
Note: Three wells need to be filled per subset (unstained, isotype mixture, and the antibody mixture).
Wash 1× with 100 μL of PBA.
Prepare three different antibody (Ab) combinations (every Ab mixture identifies one of the DC subsets) with the antibodies indicated in Table 1. The dilutions of the antibodies are made with PBA.
Table 1. Antibody mixtures to measure purity of cDC2, pDC, and cDC1 populations
Population Target Label Dilution cDC2 purity CD20 FITC 1/25 CD1c PE 1/50 CD14 PercP 1/25 CD11c APC 1/50 pDC purity BDCA2 PE 1/25 CD14 PercP 1/25 CD123 APC 1/25 CD20 FITC 1/25 cDC1 purity CD1c PE 1/50 CD14 PercP 1/25 CD141 APC 1/100 Add 25 μL of antibody mixture to the appropriate wells.
Incubate for 20 min at 4 °C in the dark.
Wash twice with 100 μL of PBA.
Resuspend in 100 μL of PBA.
Measure using a flow cytometer.
Data analysis
In the following section, examples of gating strategies for the purity assessment of the isolated DC subsets are shown after the samples were analyzed by flow cytometry.
Note: Flow cytometry data were obtained in .fcs files and uploaded to FlowJo.
For the three DC subsets, identify viable cells on FSC vs. SSC gating (Figure 4A). To evaluate the purity of the viable cDC1s, gate the cells that are negative for CD14 and CD1c (Figure 4B) and gate on CD141, the marker for cDC1s (Figure 4C).
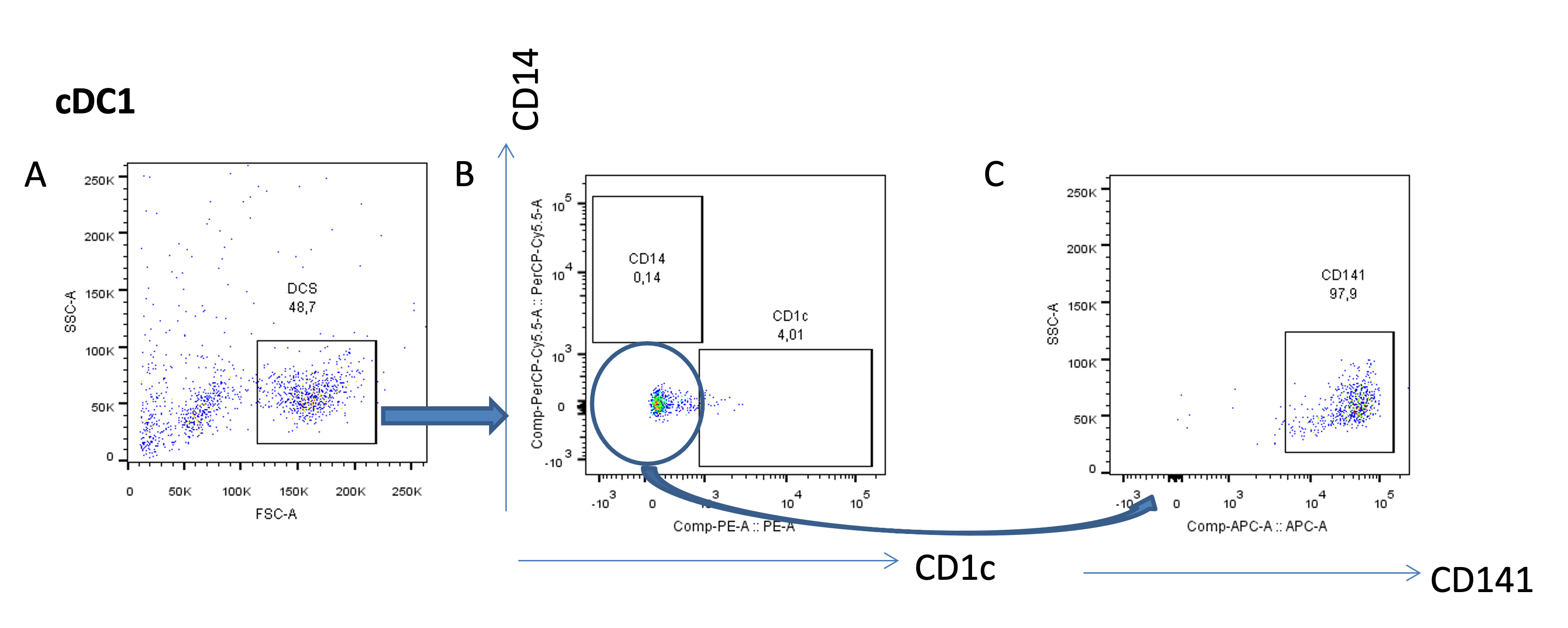
Figure 4. Purity analysis of the cDC1 subset
To evaluate the purity of the viable cDC2s (Figure 5A), gate the cells that are negative for CD14 and CD20 (Figure 5B), and gate for CD11c and CD1c double-positive cells, markers for cDC2s (Figure 5C).
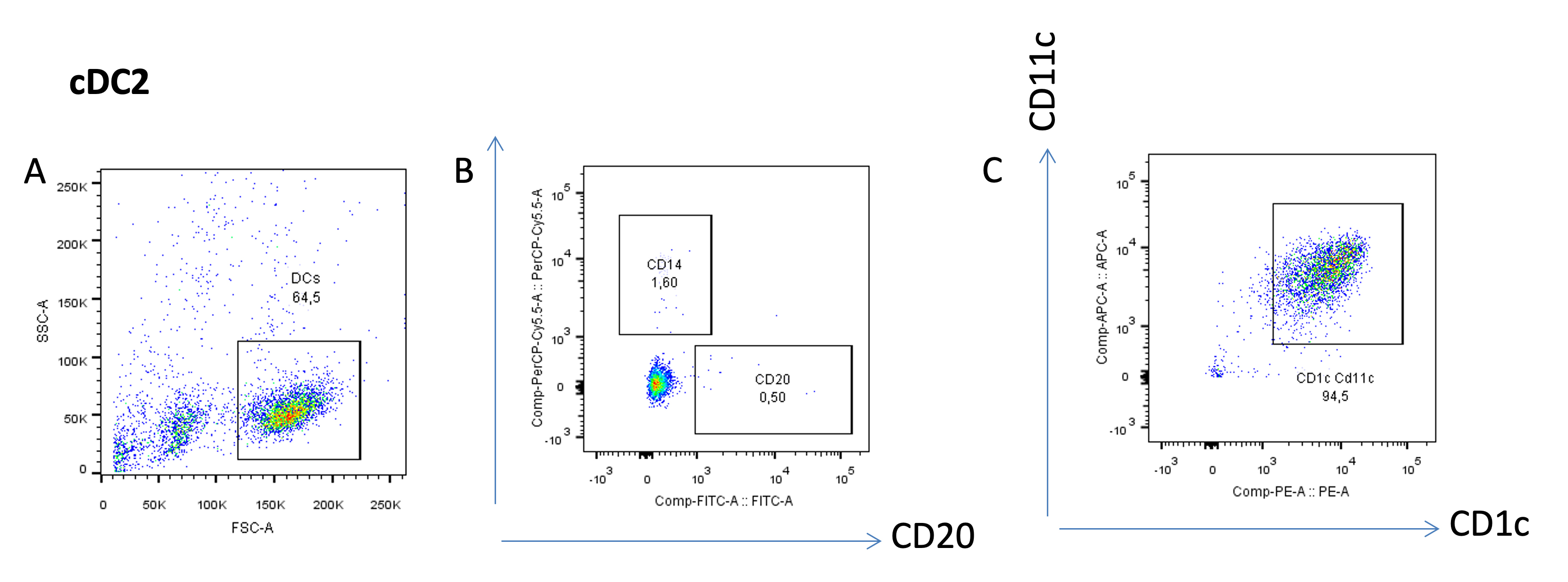
Figure 5. Purity analysis of the cDC2 subset
To evaluate the purity of the viable pDCs (Figure 6A), gate the cells that are negative for CD14 (SSC vs. CD14 plot, Figure 6B), and gate for CD123 and BDCA2 double-positive cells, markers for pDCs (Figure 6C).
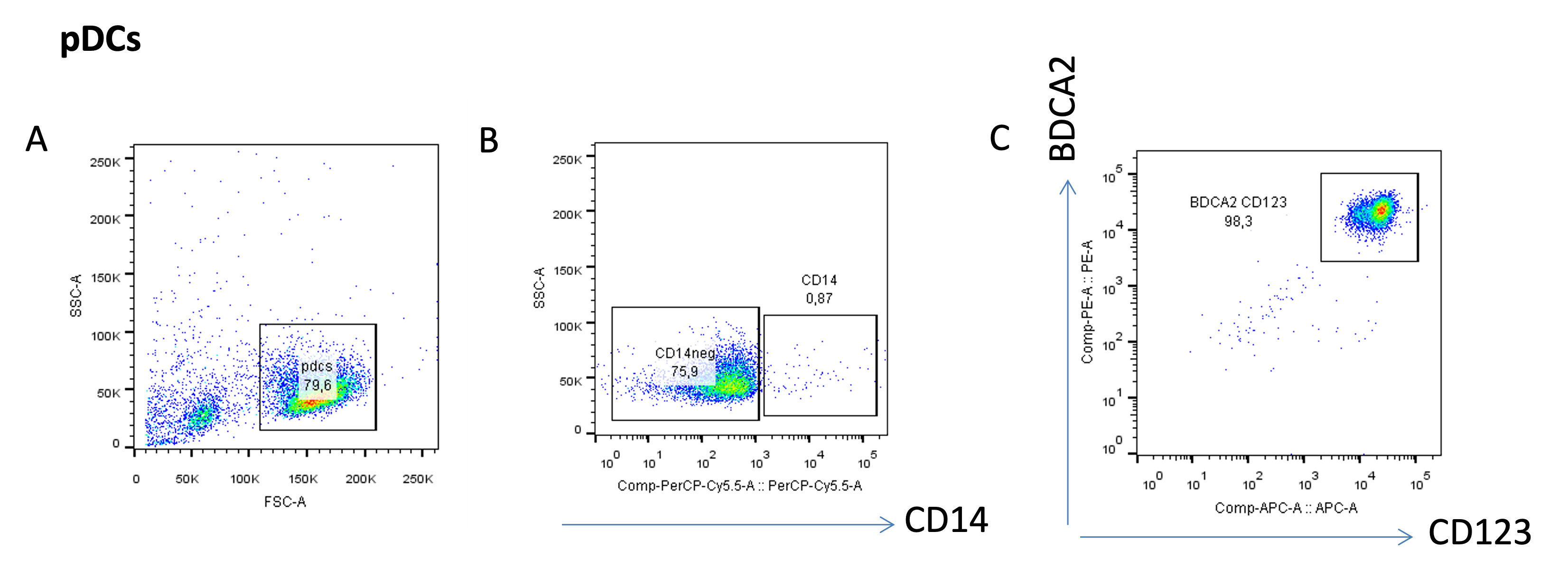
Figure 6. Purity analysis of the pDC subset
Notes
In Table 2 we provide data regarding the reproducibility and variability of the DC isolation protocol. Data come from the percentage of pure cells obtained after analysis by flow cytometry. This data corresponds to three independently performed experiments (three different donors).
Table 2. Purity of DC subsets (n = 3)
| Donor/independent experiment | Purity of cDC1s (%) | Purity of cDC2s (%) | Purity of pDCs (%) |
| Donor A | 87.5 | 94.5 | 98.3 |
| Donor B | 91.3 | 99.1 | 98.5 |
| Donor C | 99.2 | 93.2 | 99 |
Recipes
Washing buffer
2 mM EDTA and 1% bovine serum albumin (BSA) in PBS.
ACK lysis buffer
Use Milli-Q water and dissolve the following salts:
0.15 M ammonium chloride (mw: 53.49 g/mol)
0.01 M potassium bicarbonate (mw: 100.12 g/mol)
0.0001 M disodium EDTA (mw: 372.24 g/mol)
PBA buffer
Prepare by adding the reagents below to PBS. The storage of PBA is at 4 °C.
1% BSA
0.05% sodium azide (needs to be dissolved in Milli-Q water)
Validation of protocol
This protocol or parts of it has been used and validated in the following research article(s):
Le Gall et al. (2022). Efficient targeting of NY-ESO-1 tumor antigen to human cDC1s by lymphotactin results in cross-presentation and antigen-specific T cell expansion. J. Immunother Cancer (Figure 3, panel A, B, C).
General notes and troubleshooting
Troubleshooting
This protocol has limitations. To isolate human DC subsets following this protocol, there is a need for high cell numbers (PBMCs) in the starting material. Aphaeresis material instead of a buffy coat is therefore recommended. Another limitation are the high costs for the MultiMACS separator. This could be replaced by the use of single columns and magnets. However, the isolation time will increase considerably.
Acknowledgments
This work was supported by Health Holland grant DC4Balance (LSHM18056-SGF).
Competing interests
Miltenyi Biotec GmbH was as part of the public-private DC4Balance consortium. All reagents purchased to perform these experiments and the MultiMACS were bought by Radboud University Medical Center.
Ethical considerations
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donor blood (Sanquin, the Netherlands) through density centrifugation using Lymphoprep (Axis-Shield).
References
- van Beek, J. J., Flórez-Grau, G., Gorris, M. A., Mathan, T. S., Schreibelt, G., Bol, K. F., Textor, J. and de Vries, I. J. M. (2020). Human pDCs Are Superior to cDC2s in Attracting Cytolytic Lymphocytes in Melanoma Patients Receiving DC Vaccination. Cell Rep. 30(4): 1027–1038.e4.
- Collin, M. and Bigley, V. (2018). Human dendritic cell subsets: an update. Immunology 154(1): 3–20.
- Corkum, C. P., Ings, D. P., Burgess, C., Karwowska, S., Kroll, W. and Michalak, T. I. (2015). Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunology 16(1): e1186/s12865-015-0113-0.
- Gonzalez, H., Hagerling, C. and Werb, Z. (2018). Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 32: 1267–1284.
- Schreibelt, G., Klinkenberg, L. J. J., Cruz, L. J., Tacken, P. J., Tel, J., Kreutz, M., Adema, G. J., Brown, G. D., Figdor, C. G., de Vries, I. J. M., et al. (2012). The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-)presentation by human blood BDCA3+ myeloid dendritic cells. Blood 119(10): 2284–2292.
- Wang, Y., Xiang, Y., Xin, V. W., Wang, X. W., Peng, X. C., Liu, X. Q., Wang, D., Li, N., Cheng, J. T., Lyv, Y. N., et al. (2020). Dendritic cell biology and its role in tumor immunotherapy. J. Hematol. Oncol. 13(1): e1186/s13045-020-00939-6.
Article Information
Publication history
Accepted: Feb 20, 2023
Published: Oct 20, 2023
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Flórez-Grau, G., Cuenca Escalona, J., Lacasta-Mambo, H., Roelofs, D., Bödder, J., Beuk, R., Schreibelt, G. and de Vries, I. J. M. (2023). Human Dendritic Cell Subset Isolation by Magnetic Bead Sorting: A Protocol to Efficiently Obtain Pure Populations. Bio-protocol 13(20): e4851. DOI: 10.21769/BioProtoc.4851.
Category
Immunology > Immune cell isolation > Antigen-presenting cell
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







