- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Epidermal Growth Factor (EGF) Receptor Endocytosis Assay in A549 Cells
Published: Vol 3, Iss 15, Aug 5, 2013 DOI: 10.21769/BioProtoc.847 Views: 18150
Reviewed by: Lin FangFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Assessment of Human Dendritic Cell Antigen Uptake by Flow Cytometry
Ana Luque [...] Josep M. Aran
Nov 20, 2013 17117 Views
Methods to Quantify the Dynamic Recycling of Plasma Membrane Channels
Rawad Hodeify and Khaled Machaca
Sep 5, 2023 510 Views
Protein Level Quantification Across Fluorescence-based Platforms
Hector Romero [...] M. Cristina Cardoso
Oct 5, 2023 539 Views
Abstract
The following endocytosis assay has been optimized to assess EGF-stimulated EGFR endocytosis; but could be modified to assess other ligand-stimulated endocytosis of plasma membrane receptors (for which fluorochrome-conjugated ligands are available to track their receptor internalization). In brief, cells are treated with fluorescent EGF at 4 °C to allow binding to the receptor, but not internalization; then, endocytosis is allowed at 37 °C for different timepoints. For the setting up of this protocol we are really indebted to Dr. Letizia Lanzetti (Lanzetti et al., 2000).
Keywords: InternalizationMaterials and Reagents
- A549 non-small lung carcinoma cells (ATCC)
- D-MEM medium (Sigma-Aldrich)
- 10% serum RPMI medium (Sigma-Aldrich)
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A3912 )
- Fluorescent EGF-555 (Life Technologies, InvitrogenTM, catalog number: E35350 )
- 100% Acetic acid
- 5 M NaCl solution
- Paraformaldehyde (Fluka)
- Phosphate buffer saline (PBS)
- Coverslip-Slide Mounting solution (Fluoromonut, Southern Biotech)
- 4',6-diamidino-2-phenylindole (DAPI) (F. Hoffmann-La Roche)
- Saponin (Sigma-Aldrich, catalog number: S4521 )
- Acid wash (see Recipes)
- 4% PAF in PBS (see Recipes)
Equipment
- Round glass cover slips
- 6-well and 24-well dishes for mammalian cell culture (Corning, Costar®)
- Lipofectamine2000 (Life Technologies, InvitrogenTM)
- Leica TCS SP2 AOBS confocal laser-scanning microscope (Leica Microsystems)
- 4 °C refrigerator
- Cell culture incubator humidified 5% CO2 atmosphere
- pH-meter
Software
- ImageJ software (free download from http://rsb.info.nih.gov/ij/)
Procedure
Day 1
Seed the appropriate number of cells (see Note 2) on top of glass coverslips placed (one per well) in 24-well cell culture dishes. Alternatively, if it is required to transfect cells prior to performing the endocytosis assay, seed cells on top of 5-6 coverslips placed in each well of 6-well culture dishes. You should prepare at least one independent multi-well dish per each time point in your experiment, plus two for controls (e.g. four dishes for an experiment with control cells plus two different times of endocytosis).
Day 2 (optional)
Cell DNA-transfection with Lipofectamine2000, according to manufacturer’s protocol. On the day of the endocytosis assay, transfer the coverslips with cells into 24-well cell culture dishes. Transfection could be needed when you need to modulate proteins putatively involved in the internalization of the receptor.
Day 3-4
Endocytosis assay (or Day 2-3, if no cell transfection needed prior to the assay), by the following steps (see Figure 1 for the protocol scheme).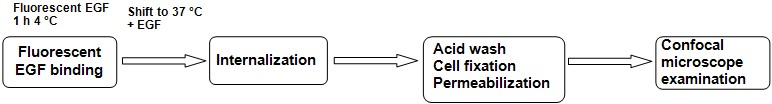
Figure 1. Schematic representation of experimental flow
- Remove cell culture medium and incubate cells with 400-500 μl of serum-free DMEM medium containing 0.3% BSA for at least 1 h (extendable up to 3 h) at 37 °C in cell culture incubator, in order to deprive cells of serum-contained stimuli.
- Replace cell medium with the appropriate dilution of the fluorescent ligand EGF (2 ng/ml) into 400 μl of DMEM 0.3% BSA (per well).
- Incubate the cells on ice for 1 h, in the dark (by covering the dish with aluminium foil).
- Wash two multi-well dishes with ice-cold PBS three times and leave them on ice (control condition = ligand binding without endocytosis, at 4 °C binding ligand receptor is allowed, while internalization is blocked).
- On one of the two dishes above, perform an acidic wash (to remove non-internalized ligand) by incubating the cells with an acetic acid buffer on ice for 6 min. Rinse with ice-cold PBS three times and leave on ice.
- Transfer the other cell culture dishes at 37 °C to elicit endocytosis for different periods (optimal time points need to be set for the specific cellular model and ligand stimulation; in our experience, for example, 12 and 25 min for EGF 2 ng/ml to A549 cells).
- At the end of each incubation, block endocytosis by moving dishes on ice.
- Wash 3 times with ice-cold PBS and proceed with acid wash on ice as described in step 5 (to detach all the ligand bound to the receptor at the plasma membrane and allow detection of internalized ligand only).
- After acidic wash, rinse each well 3 times with cold PBS.
- Fix cells on every coverslip by incubation with 4% PFA in PBS for 10 min at room temperature and then wash 3 times with PBS. Fixed cells may be stored for several days in the refrigerator at this stage.
- Permeabilize cell membranes by incubating fixed cells on coverslips with a solution of 0.01% saponin in PBS 1% BSA for 5-10 min on ice (in alternative, you may use 0.1% Triton in PBS 1% BSA).
- Wash permeabilized cells three times with PBS.
- (Optional) If Immunofluorescence analysis (IFA) is planned, non-specific binding may be blocked by incubation with PBS 1% BSA for 30 min, followed by immunostaining with specific antibodies.
- Counterstain cell nuclei by incubating with DAPI 5 min at room temperature (a dilution 1:10,000 of a stock of 1 μg/μl).
- Finally, wash once coverslips in PBS, then rinse with water and fix them inverted on microscope glass slides by including a mounting medium.
- Analysis of fluorescent signals should be done with a confocal microscope with a 63x magnification, such as Leica TCS SP2 AOBS confocal laser-scanning microscope (see Figure 2 for representative images).
- Quantify endocytosed fluorescent signal acquired with ImageJ software.
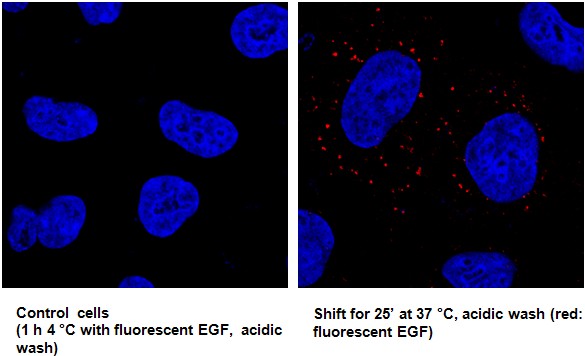
Figure 2. Representative pictures of endocytosis assay on A549. On the left, control cells treated with fluorescent EGF 1 h at 4 °C and subjected to acidic wash to clear out all EGF bound to plasmamembrane; no fluorescent signal from the plasmamembrane. On the right, cells treated with fluorescent EGF 1 h at 4 °C, subjected to acidic wash and then shifted to 37 °C (internalization); fluorescent signal from internalized EGF.
Notes
- I settled this protocol on A549 cells, other cell lines may require optimization of experimental conditions (such as different timing of endocytosis induction at 37 °C). A549 should be grown in 10% serum RPMI medium.
- Grow cells on coverslips in their optimal culture medium to reach 50-80% confluency on the day of the assay. Remember to have in separate multiwell dishes the coverslips that will have to undergo the different incubation times or control conditions.
- In the meanwhile of step 2 (cell incubation with 0.3% BSA solution), prepare fluorochrome-conjugated ligand solution at the desired final concentration in DMEM 0.3% BSA, considering 400-500 μl per each well (in 24-well dishes). Remember to keep ligand-containing solutions always in the dark (covered with aluminium foil).
- Put a plastic box (large enough to contain all multiwell dishes) with one inch of water on the bottom to warm up at 37 °C in the cell incubator that you will use for the assay. This will serve to speed up the temperature change between 4 °C and 37 °C to allow accurate timing for endocytosis in the different conditions.
- Pay attention not to put the acidic wash solution directly on cells to avoid causing their detachment, but instead gently pour it on the side of the wells.
- Do not exceed 6 min incubation with acidic wash. Acidic wash for coverslips always kept on ice, will provide the negative control of the experiments: In fact, after acidic wash, you should not detect any fluorescent signal from the ligand associated with the cells. On the other hand, there should still be strong signal on PBS-washed cells not subjected to acidic wash, indicating that ligand binding to the receptor indeed occurred.
- It is good rule to acquire at least 50 different microscopic fields per each condition, from at least three independent experiments (applying constant detection parameters at the confocal microscope). Mean fluorescence intensity of the fields should then be quantified from image files (e.g. by ImageJ software).
Recipes
- Acid wash
0.2 M acetic acid
0.5 M NaCl (pH 2.8)
For 100 ml
1.2 ml 100% acetic acid
10 ml 5 M NaCl
88.8 ml H2O - 4% PFA in PBS
4 g paraformaldehyde in 100 ml PBS
Heat PBS (shaking) for 30 min until it reaches 60 °C
Add paraformaldehyde
Clarify by dropwise addiction of 5 M NaOH
Cool down temperature while shaking
Adjust pH at 7.4 (with 5 M NaOH/6 N HCl)
Bring to final volume with PBS
Shake again the solution
Aliquote
Keep at -20 °C
Acknowledgments
This protocol was initially developed for the study Rizzolio et al. (2012). Previous reports analyzing receptors internalization that have been used as reference to set up this procedure include: Lanzetti et al. (2000); Sawamiphak et al. (2010) and Popovic et al. (2012). Correlated research activity in the authors’ lab was funded by the University of Torino-Compagnia di San Paolo, Grant ORTO11RKTW (RETHE). The authors are indebted to Dr. Letizia Lanzetti (University of Torino) for her expert support in setting up this protocol.
References
- Lanzetti, L., Rybin, V., Malabarba, M. G., Christoforidis, S., Scita, G., Zerial, M. and Di Fiore, P. P. (2000). The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408(6810): 374-377.
- Popovic, D., Akutsu, M., Novak, I., Harper, J. W., Behrends, C. and Dikic, I. (2012). Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 32(9): 1733-1744.
- Rizzolio, S., Rabinowicz, N., Rainero, E., Lanzetti, L., Serini, G., Norman, J., Neufeld, G. and Tamagnone, L. (2012). Neuropilin-1-dependent regulation of EGF-receptor signaling. Cancer Res 72(22): 5801-5811.
- Sawamiphak, S., Seidel, S., Essmann, C. L., Wilkinson, G. A., Pitulescu, M. E., Acker, T. and Acker-Palmer, A. (2010). Ephrin-B2 regulates VEGFR2 function in developmental and tumour angiogenesis. Nature 465(7297): 487-491.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rizzolio, S. and Tamagnone, L. (2013). Epidermal Growth Factor (EGF) Receptor Endocytosis Assay in A549 Cells. Bio-protocol 3(15): e847. DOI: 10.21769/BioProtoc.847.
Category
Cell Biology > Cell imaging > Fluorescence
Cell Biology > Cell-based analysis > Cytosis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







